Fecal Source Tracking in A Wastewater Treatment and Reclamation System Using Multiple Waterborne Gastroenteritis Viruses
Abstract
:1. Introduction
2. Results
2.1. Occurrence of Viral Genes in Wastewater Samples
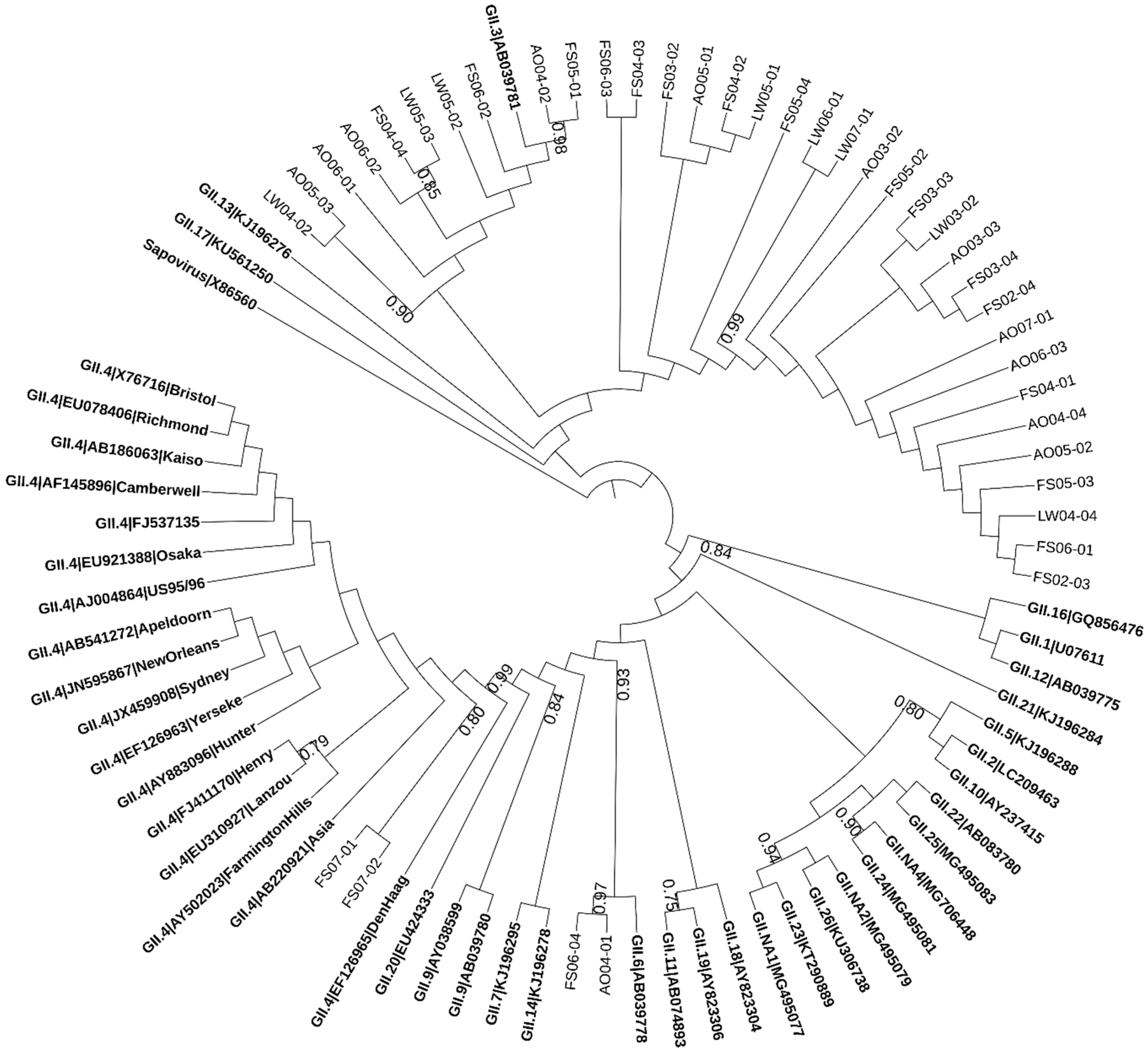
2.2. Phylogenetic Analysis of Norovirus
2.3. Molecular Detection and Characterization of Rotavirus
3. Discussion
4. Materials and Methods
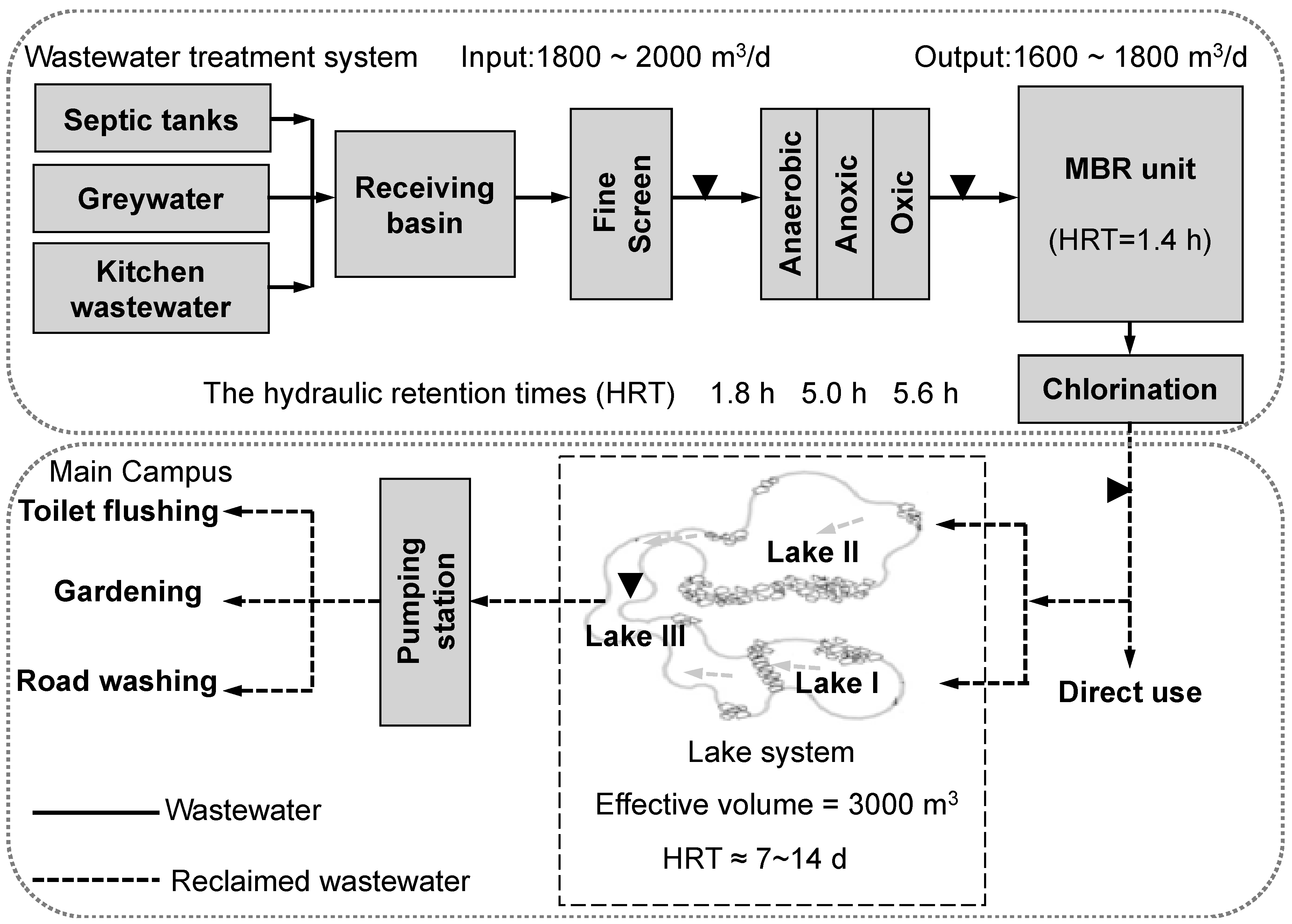
4.1. Sample Collection
4.2. Recovery of Viral Particles and Nucleic Acid Extraction
4.3. Molecular Detection and Characterization of Enteric Viruses
4.4. Nucleotide Sequencing and Phylogenetic Analysis
4.5. Nucleotide Sequence Accession Numbers
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Angelakis, A.N.; Asano, T.; Bahri, A.; Jimenez, B.E.; Tchobanoglous, G. Water reuse: From ancient to modern times and the future. Front. Environ. Sci. 2018, 6, 26. [Google Scholar] [CrossRef]
- Zhang, C.-M.; Wang, X.-C. Distribution of enteric pathogens in wastewater secondary effluent and safety analysis for urban water reuse. Hum. Ecol. Risk Assess. 2014, 20, 797–806. [Google Scholar] [CrossRef]
- Sano, D.; Amarasiri, M.; Hata, A.; Watanabe, T.; Katayama, H. Risk management of viral infectious diseases in wastewater reclamation and reuse: Review. Environ. Int. 2016, 91, 220–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bofill-Mas, S.; Rusiñol, M.; Fernandez-Cassi, X.; Carratalà, A.; Hundesa, A.; Girones, R. Quantification of human and animal viruses to differentiate the origin of the fecal contamination present in environmental samples. BioMed Res. Int. 2013, 2013, 192089. [Google Scholar] [CrossRef]
- García-Aljaro, C.; Blanch, A.R.; Campos, C.; Jofre, J.; Lucena, F. Pathogens, faecal indicators and human-specific microbial source-tracking markers in sewage. J. Appl. Microbiol. 2019, 126, 701–717. [Google Scholar] [CrossRef] [PubMed]
- Field, K.G.; Samadpour, M. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res. 2007, 41, 3517–3538. [Google Scholar] [CrossRef]
- O’Mullan, G.D.; Dueker, M.E.; Juhl, A.R. Challenges to managing microbial fecal pollution in coastal environments: Extra-enteric ecology and microbial exchange among water, sediment, and air. Curr. Pollut. Rep. 2017, 3, 1–16. [Google Scholar]
- García-Aljaro, C.; Ballesté, E.; Muniesa, M.; Jofre, J. Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb. Biotechnol. 2017, 10, 1775–1780. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.A.; Vega, A.A.; Norman, H.M.; Ohaeri, M.; Levi, K.; Dinsdale, E.A.; Cinek, O.; Aziz, R.K.; McNair, K.; Barr, J.J.; et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 2019, 527796. [Google Scholar] [CrossRef]
- Mclellan, S.L.; Eren, A.M. Discovering new indicators of fecal pollution. Trends Microbiol. 2014, 22, 697–706. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.; Fong, T.-T.; Bibby, K.; Molina, M. Application of enteric viruses for fecal pollution source tracking in environmental waters. Environ. Int. 2012, 45, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Boehm, A.B.; Sassoubre, L.M.; Vijayavel, K.; Stewart, J.R.; Fong, T.-T.; Caprais, M.-P.; Converse, R.R.; Diston, D.; Ebdon, J.; et al. Performance of viruses and bacteriophages for fecal source determination in a multi-laboratory, comparative study. Water Res. 2013, 47, 6929–6943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rachmadi, A.T.; Torrey, J.R.; Kitajima, M. Human polyomavirus: Advantages and limitations as a human-specific viral marker in aquatic environments. Water Res. 2016, 105, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Bofill-Mas, S.; Albinana-Gimenez, N.; Clemente-Casares, P.; Hundesa, A.; Rodriguez-Manzano, J.; Allard, A.; Calvo, M.; Girones, R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 2006, 72, 7894–7896. [Google Scholar] [CrossRef] [PubMed]
- Amarasiri, M.; Kawai, H.; Kitajima, M.; Okabe, S.; Sano, D. Specific interactions of rotavirus HAL1166 with Enterobacter cloacae SENG-6 and their contribution on rotavirus HAL1166 removal. Water Sci. Technol. 2019, 79, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Amarasiri, M.; Sano, D. Specific interactions between human norovirus and environmental matrices: Effects on the virus ecology. Viruses 2019, 11, 224. [Google Scholar] [CrossRef]
- Miura, T.; Okabe, S.; Nakahara, Y.; Sano, D. Removal properties of human enteric viruses in a pilot-scale membrane bioreactor (MBR) process. Water Res. 2015, 75, 282–291. [Google Scholar] [CrossRef] [Green Version]
- Amarasiri, M.; Kitajima, M.; Nguyen, T.H.; Okabe, S.; Sano, D. Bacteriophage removal efficiency as a validation and operational monitoring tool for virus reduction in wastewater reclamation: Review. Water Res. 2017, 121, 258–269. [Google Scholar] [CrossRef]
- Ottoson, J.; Hansen, A.; Björlenius, B.; Norder, H.; Stenström, T.A. Removal of viruses, parasitic protozoa and microbial indicators in conventional and membrane processes in a wastewater pilot plant. Water Res. 2006, 40, 1449–1457. [Google Scholar] [CrossRef]
- Marti, E.; Monclús, H.; Jofre, J.; Rodriguez-roda, I.; Comas, J.; Luis, J. Removal of microbial indicators from municipal wastewater by a membrane bioreactor (MBR). Bioresour. Technol. 2011, 102, 5004–5009. [Google Scholar] [CrossRef]
- World Health Organization. Guidelines for Drinking Water Quality: Fourth Edition Incorporating the First Addendum, 4th ed.; World Health Organization: Geneva, Switzerland, 2017; ISBN 9789241549950. [Google Scholar]
- Satter, S.M.; Aliabadi, N.; Gastañaduy, P.A.; Haque, W.; Mamun, A.; Flora, M.S.; Zaman, K.; Rahman, M.; Heffelfinger, J.D.; Luby, S.P.; et al. An update from hospital-based surveillance for rotavirus gastroenteritis among young children in Bangladesh, July 2012 to June 2017. Vaccine 2018, 36, 7811–7815. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Chughtai, A.A.; Gao, Z.; Yan, H.; Chen, Y.; Liu, B.; Huo, D.; Jia, L.; Wang, Q.; MacIntyre, C.R. Prevalence and genotypes of group A rotavirus among outpatient children under five years old with diarrhea in Beijing, China, 2011–2016. BMC Infect. Dis. 2018, 18, 497. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-D.; Unno, H. The roles of microbes in the removal and inactivation of viruses in a biological wastewater treatment system. Water Sci. Technol. 1996, 33, 243–250. [Google Scholar] [CrossRef]
- Amarasiri, M.; Hashiba, S.; Miura, T.; Nakagomi, T.; Nakagomi, O.; Ishii, S.; Okabe, S.; Sano, D. Bacterial histo-blood group antigens contributing to genotype-dependent removal of human noroviruses with a microfiltration membrane. Water Res. 2016, 95, 383–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, B.W.; Kitajima, M.; Campillo, M.E.; Gerba, C.P.; Pepper, I.L. Virus reduction during advanced Bardenpho and conventional wastewater treatment processes. Environ. Sci. Technol. 2016, 50, 9524–9532. [Google Scholar] [CrossRef] [PubMed]
- Arraj, A.; Bohatier, J.; Laveran, H.; Traore, O. Comparison of bacteriophage and enteric virus removal in pilot scale activated sludge plants. J. Appl. Microbiol. 2005, 98, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-M.; Xu, L.-M.; Xu, P.-C.; Wang, X.C. Elimination of viruses from domestic wastewater: Requirements and technologies. World J. Microbiol. Biotechnol. 2016, 32, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Villabruna, N.; Koopmans, M.P.G.; De Graaf, M. Animals as reservoir for human norovirus. Viruses 2019, 11, 478. [Google Scholar] [CrossRef] [PubMed]
- Summa, M.; Henttonen, H.; Maunula, L. Human noroviruses in the faeces of wild birds and rodents—New potential transmission routes. Zoonoses Public Health 2018, 65, 512–518. [Google Scholar] [CrossRef]
- Uhrbrand, K.; Schultz, A.C.; Madsen, A.M. Exposure to airborne noroviruses and other bioaerosol components at a wastewater treatment plant in Denmark. Food Environ. Virol. 2011, 3, 130–137. [Google Scholar] [CrossRef]
- Dueker, M.E.; O’Mullan, G.D.; Martinez, J.M.; Juhl, A.R.; Weathers, K.C. Onshore wind speed modulates Microbial aerosols along an urban waterfront. Atmosphere 2017, 8, 215. [Google Scholar] [CrossRef]
- Ueki, Y.; Sano, D.; Watanabe, T.; Akiyama, K.; Omura, T. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oysters. Water Res. 2005, 39, 4271–4280. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, T.; Kojima, S.; Shinohara, M.; Uchida, K.; Fukushi, S.; Hoshino, F.B.; Takeda, N.; Katayama, K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003, 41, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Amarasiri, M.; Kitajima, M.; Miyamura, A.; Santos, R.; Monteiro, S.; Miura, T.; Kazama, S.; Okabe, S.; Sano, D. Reverse transcription-quantitative PCR assays for genotype-specific detection of human noroviruses in clinical and environmental samples. Int. J. Hyg. Environ. Health 2018, 221, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Eftim, S.E.; Hong, T.; Soller, J.; Boehm, A.; Warren, I.; Ichida, A.; Nappier, S.P. Occurrence of norovirus in raw sewage—A systematic literature review and meta-analysis. Water Res. 2017, 111, 366–374. [Google Scholar] [CrossRef]
- Wyn-Jones, A.P.; Carducci, A.; Cook, N.; D’Agostino, M.; Divizia, M.; Fleischer, J.; Gantzer, C.; Gawler, A.; Girones, R.; Höller, C.; et al. Surveillance of adenoviruses and noroviruses in European recreational waters. Water Res. 2011, 45, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Haramoto, E.; Oguma, K.; Yamashita, H.; Tajima, A.; Nakajima, H.; Ohgaki, S. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 2008, 42, 1441–1448. [Google Scholar] [CrossRef]
- Da Silva, A.K.; Le Saux, J.-C.; Parnaudeau, S.; Pommepuy, M.; Elimelech, M.; Le Guyader, F.S. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: Different behaviors of genogroups I and II. Appl. Environ. Microbiol. 2007, 73, 7891–7897. [Google Scholar] [CrossRef]
- Da Silva, A.K.; Le Guyader, F.S.; Le Saux, J.C.; Pommepuy, M.; Montgomery, M.A.; Elimelech, M. Norovirus removal and particle association in a waste stabilization pond. Environ. Sci. Technol. 2008, 42, 9151–9157. [Google Scholar] [CrossRef]
- Grassi, T.; Bagordo, F.; Idolo, A.; Lugoli, F.; Gabutti, G.; De Donno, A. Rotavirus detection in environmental water samples by tangential flow ultrafiltration and RT-nested PCR. Environ. Monit. Assess. 2010, 164, 199–205. [Google Scholar] [CrossRef]
- Kiulia, N.; Hofstra, N.; Vermeulen, L.; Obara, M.; Medema, G.; Rose, J. Global occurrence and emission of rotaviruses to surface waters. Pathogens 2015, 4, 229–255. [Google Scholar] [CrossRef] [PubMed]
- Baggi, F.; Peduzzi, R. Genotyping of rotaviruses in environmental water and stool samples in Southern Switzerland by nucleotide sequence analysis of 189 base pairs at the 5′ end of the VP7 gene. J. Clin. Microbiol. 2000, 38, 3681–3685. [Google Scholar] [PubMed]
- Tree, J.A.; Adams, M.R.; Lees, D.N. Chlorination of indicator bacteria and viruses in primary sewage effluent. Appl. Environ. Microbiol. 2003, 69, 2038–2043. [Google Scholar] [CrossRef] [PubMed]
- Albinana-Gimenez, N.; Miagostovich, M.P.; Calgua, B.; Huguet, J.M.; Matia, L.; Girones, R. Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. Water Res. 2009, 43, 2011–2019. [Google Scholar] [CrossRef]
- Haramoto, E.; Fujino, S.; Otagiri, M. Distinct behaviors of infectious F-specific RNA coliphage genogroups at a wastewater treatment plant. Sci. Total Environ. 2015, 520, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Kazama, S.; Masago, Y.; Tohma, K.; Souma, N.; Imagawa, T.; Suzuki, A.; Liu, X.; Saito, M.; Oshitani, H.; Omura, T. Temporal dynamics of norovirus determined through monitoring of municipal wastewater by pyrosequencing and virological surveillance of gastroenteritis cases. Water Res. 2016, 92, 244–253. [Google Scholar] [CrossRef] [Green Version]
- Kazama, S.; Miura, T.; Masago, Y.; Konta, Y.; Tohma, K.; Manaka, T.; Liu, X.; Nakayama, D.; Tanno, T.; Saito, M.; et al. Environmental Surveillance of norovirus genogroups I and II for sensitive detection of epidemic variants. Appl. Environ. Microbiol. 2017, 83, e03406-16. [Google Scholar] [CrossRef] [PubMed]
- Haramoto, E.; Kitajima, M.; Hata, A.; Torrey, J.R.; Masago, Y.; Sano, D.; Katayama, H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018, 135, 168–186. [Google Scholar] [CrossRef]
- Mijatovic-rustempasic, S.; Esona, M.D.; Williams, A.L.; Bowen, M.D. Sensitive and specific nested PCR assay for detection of rotavirus A in samples with a low viral load. J. Virol. Methods 2016, 236, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Lew, A.M.; Marshall, V.M.; Kemp, D.J. Affinity selection of polymerase chain reaction products by DNA-binding proteins. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1993; Volume 218, pp. 526–534. [Google Scholar]
- Ma, X.Y.; Wang, X.C.; Wang, D.; Ngo, H.H.; Zhang, Q.; Wang, Y.; Dai, D. Function of a landscape lake in the reduction of biotoxicity related to trace organic chemicals from reclaimed water. J. Hazard. Mater. 2016, 318, 663–670. [Google Scholar] [CrossRef]
- Gao, T.; Chen, R.; Wang, X.; Hao, H.; Li, Y.; Zhou, J.; Zhang, L. Application of disease burden to quantitative assessment of health hazards for a decentralized water reuse system. Sci. Total Environ. 2016, 551–552, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.D.; Metcalf, T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988, 54, 1983–1988. [Google Scholar] [PubMed]
- Ji, Z.; Wang, X.; Zhang, C.; Miura, T.; Sano, D.; Funamizu, N.; Okabe, S. Occurrence of hand-foot-and-mouth disease pathogens in domestic sewage and secondary effluent in Xi’an, China. Microbes Environ. 2012, 27, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Shimasaki, A.; Ohgaki, S. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 2002, 68, 1033–1039. [Google Scholar] [CrossRef]
- O’Neill, H.J.; McCaughey, C.; Coyle, P.V.; Wyatt, D.E.; Mitchell, F. Clinical utility of nested multiplex RT-PCR for group F adenovirus, rotavirus and norwalk-like viruses in acute viral gastroenteritis in children and adults. J. Clin. Virol. 2002, 25, 335–343. [Google Scholar] [CrossRef]
- Gilgen, M.; Germann, D.; Lüthy, J.; Hübner, P. Three-step isolation method for sensitive detection of enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. Int. J. Food Microbiol. 1997, 37, 189–199. [Google Scholar] [CrossRef]
- Kojima, S.; Kageyama, T.; Fukushi, S.; Hoshino, F.B.; Shinohara, M.; Uchida, K.; Natori, K.; Takeda, N.; Katayama, K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 2002, 100, 107–114. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution (N. Y.) 1985, 39, 783–791. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]


| Virus | Sampling Locations % (Positive/Total Samples) | Total Detection Rate for Each Virus (%) | |||
|---|---|---|---|---|---|
| Mixed Raw Sewage | A2O Effluent | MBR Effluent after Disinfection | Lake Water | ||
| HuNoV GI | 67 (16/24) | 45 (11/24) | 0 (0/24) | 38 (9/24) | 38 (36/96) |
| HuNoV GII | 79 (19/24) | 50 (12/24) | 0 (0/24) | 33 (8/24) | 41 (39/96) |
| HRVs | 75 (18/24) | 29 (7/24) | 0 (0/24) | 25 (6/24) | 32 (31/96) |
| Total Detection Rate for Each Sampling Site (%) | 92 (22/24) | 71 (17/24) | 0 (0/24) | 63 (15/24) | 56 (54/96) |
| Virus | Target Gene | PCR Round | Primer | Sequence (5’-3’) a | Reference |
|---|---|---|---|---|---|
| Rotavirus | VP7(G) | 1st | RoA b | CTTTAAAAGAGAGAATTTCCGTCTG | [57,58] |
| 1st | RoB b | TGATGATCCCATTGATATCC | |||
| 2nd | RoC b | TGTATGGTATTGAATATACCAC | |||
| 2nd | RoD b | ACTGATCCTGTTGGCCAWCC | |||
| Norovirus GI | ORF1–ORF2 junction | 1st | COG1F c | CGYTGGATGCGNTTYCATGA | [34,59] |
| 1st | G1-SKR c | CCAACCCARCCATTRTACA | |||
| 2nd | G1-SKF c | CTGCCCGAATTYGTAAATGA | |||
| 2nd | G1-SKR c | CCAACCCARCCATTRTACA | |||
| Norovirus GII | ORF1–ORF2 junction | 1st | COG2F d | CARGARBCNATGTTYAGRTGGATGAG | [34,59] |
| 1st | G2-SKR e | CCRCCNGCATRHCCRTTRTACAT | |||
| 2nd | G2-SKF e | CNTGGGAGGGCGATCGCAA | |||
| 2nd | G2-SKR e | CCRCCNGCATRHCCRTTRTACAT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Z.; Wang, X.C.; Xu, L.; Zhang, C.; Rong, C.; Rachmadi, A.T.; Amarasiri, M.; Okabe, S.; Funamizu, N.; Sano, D. Fecal Source Tracking in A Wastewater Treatment and Reclamation System Using Multiple Waterborne Gastroenteritis Viruses. Pathogens 2019, 8, 170. https://doi.org/10.3390/pathogens8040170
Ji Z, Wang XC, Xu L, Zhang C, Rong C, Rachmadi AT, Amarasiri M, Okabe S, Funamizu N, Sano D. Fecal Source Tracking in A Wastewater Treatment and Reclamation System Using Multiple Waterborne Gastroenteritis Viruses. Pathogens. 2019; 8(4):170. https://doi.org/10.3390/pathogens8040170
Chicago/Turabian StyleJi, Zheng, Xiaochang C. Wang, Limei Xu, Chongmiao Zhang, Cheng Rong, Andri Taruna Rachmadi, Mohan Amarasiri, Satoshi Okabe, Naoyuki Funamizu, and Daisuke Sano. 2019. "Fecal Source Tracking in A Wastewater Treatment and Reclamation System Using Multiple Waterborne Gastroenteritis Viruses" Pathogens 8, no. 4: 170. https://doi.org/10.3390/pathogens8040170
APA StyleJi, Z., Wang, X. C., Xu, L., Zhang, C., Rong, C., Rachmadi, A. T., Amarasiri, M., Okabe, S., Funamizu, N., & Sano, D. (2019). Fecal Source Tracking in A Wastewater Treatment and Reclamation System Using Multiple Waterborne Gastroenteritis Viruses. Pathogens, 8(4), 170. https://doi.org/10.3390/pathogens8040170






