Dual Transcriptional Profile of Aspergillus flavus during Co-Culture with Listeria monocytogenes and Aflatoxin B1 Production: A Pathogen–Pathogen Interaction
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Sample Preparation
2.2. Qualitative and Quantitative Aflatoxin B1 Determination
2.3. Gene Expression Assay
2.4. Statistical Analysis
3. Results
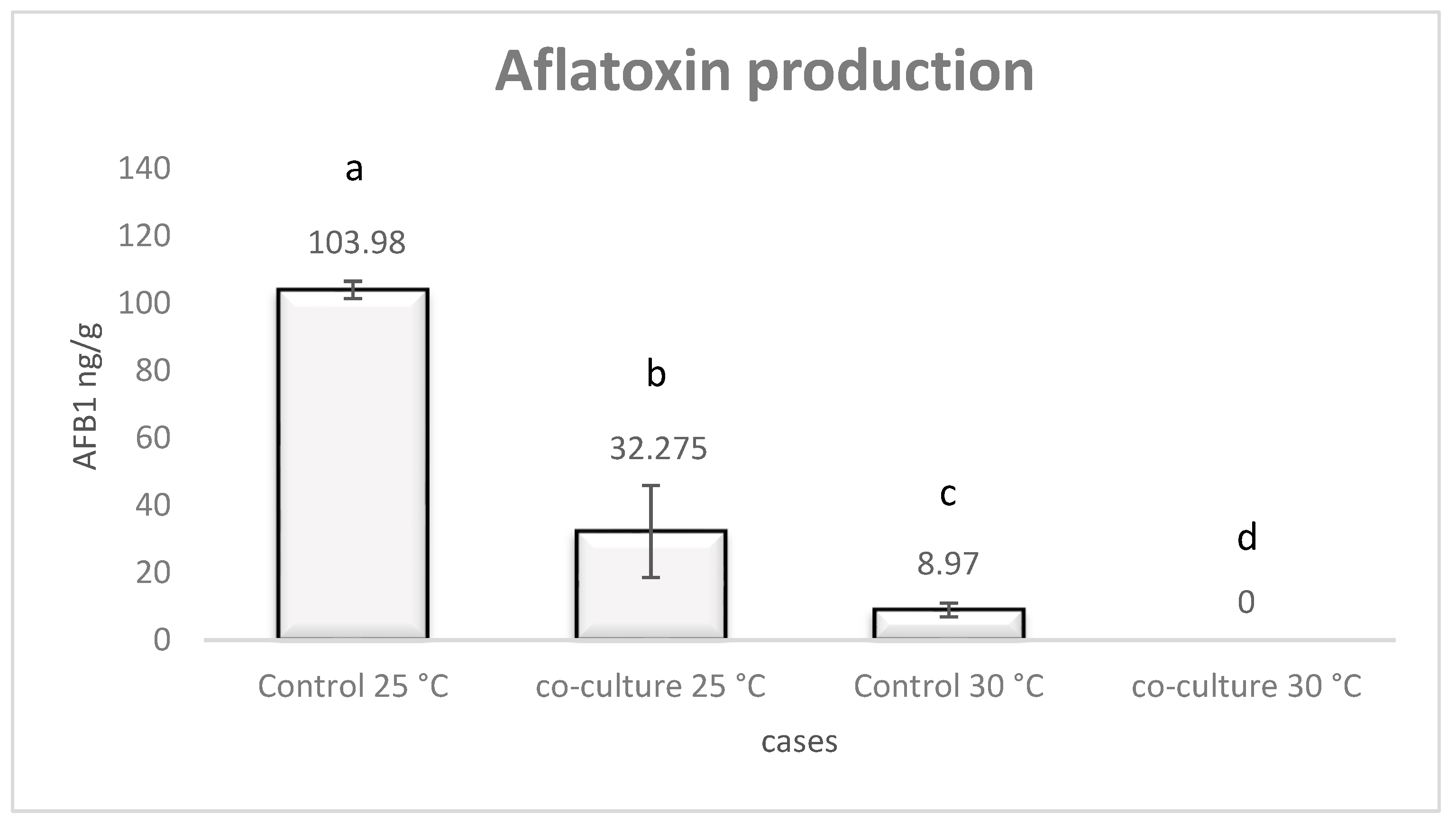
3.1. Aflatoxin B1 Production by A. flavus under Co-Culture with L. monocytogenes
3.2. Impact of Co-Culture upon Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2009, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Ravn, L.; Rasch, M.; Bruhn, J.B.; Christensen, A.B.; Givskov, M. Food spoilage—Interactions between food spoilage bacteria. Int. J. Food Microbiol. 2002, 78, 79–97. [Google Scholar] [CrossRef]
- Haruta, S.; Kato, S.; Yamamoto, K.; Igarashi, Y. Intertwined interspecies relationships: Approaches to untangle the microbial network. Environ. Microbiol. 2009, 11, 2963–2969. [Google Scholar] [CrossRef] [PubMed]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Hervé, V.; Labbé, J.; Lastovetsky, O.A.; et al. Bacterial–fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef]
- Wargo, M.J.; Hogan, D.A. Fungal—Bacterial interactions: A mixed bag of mingling microbes. Curr. Opin. Microbiol. 2006, 9, 359–364. [Google Scholar] [CrossRef]
- Casquete, R.; Benito, M.J.; Córdoba, M.D.G.; Ruiz-Moyano, S.; Martín, A. The growth and aflatoxin production of Aspergillus flavus strains on a cheese model system are influenced by physicochemical factors. J. Dairy Sci. 2017, 100, 6987–6996. [Google Scholar] [CrossRef]
- Umesha, S.; Manukumar, H.M.G.; Chandrasekhar, B.; Shivakumara, P.; Shiva Kumar, J.; Raghava, S.; Avinash, P.; Shirin, M.; Bharathi, T.R.; Rajini, S.B.; et al. Aflatoxins and food pathogens: Impact of biologically active aflatoxins and their control strategies. J. Sci. Food Agric. 2017, 97, 1698–1707. [Google Scholar] [CrossRef]
- Gómez, D.; Iguácel, L.P.; Rota, M.C.; Carramiñana, J.J.; Ariño, A.; Yangüela, J. Occurrence of Listeria monocytogenes in ready-to-eat meat products and meat processing plants in Spain. Foods 2015, 4, 271–282. [Google Scholar] [CrossRef]
- Casquete, R.; Benito, M.J.; Aranda, E.; Martín, A.; Ruiz-Moyano, S.; de Córdoba, M.G. Gene expression of Aspergillus flavus strains on a cheese model system to control aflatoxin production. J. Dairy Sci. 2019, 102, 7765–7772. [Google Scholar] [CrossRef]
- Asurmendi, P.; Barberis, C.; Dalcero, A.; Pascual, L.; Barberis, L. Survey of Aspergillus section Flavi and aflatoxin B1 in brewer’s grain used as pig feedstuff in Córdoba, Argentina. Mycotoxin Res. 2003, 29, 3–7. [Google Scholar] [CrossRef]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2013, 16, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Groopman, J.D.; Wogan, G.N. Aflatoxin and hepatocellular carcinoma. In Chemical Carcinogenesis; Penning, T.M., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 113–133. [Google Scholar]
- Wild, C.P.; Gong, Y.Y. Mycotoxins and human disease: A largely ignored global health issue. Carcinogenesis 2010, 31, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Perspect. 2010, 118, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.J.; Wałecka-Zacharska, E.; Chen, J.C.; Katarzyna, K.-P.; Devlieghere, F.; van Meervenne, E.; Osek, J.; Wieczorek, K.; Bania, J. Listeria monocytogenes—An examination of food chain factors potentially contributing to antimicrobial resistance. Food Microbiol. 2016, 54, 178–189. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Andritsos, N.D.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Listeria monocytogenes serotype prevalence and biodiversity in diverse food products. J. Food Prot. 2014, 77, 2115–2120. [Google Scholar] [CrossRef]
- Zilelidou, E.A.; Skandamis, P.N. Growth, detection and virulence of Listeria monocytogenes in the presence of other microorganisms: Microbial interactions from species to strain level. Int. J. Food Microbiol. 2018, 277, 10–25. [Google Scholar] [CrossRef]
- Mellefont, L.A.; McMeekin, T.A.; Ross, T. Effect of relative inoculum concentration on Listeria monocytogenes growth in co-culture. Int. J. Food Microbiol. 2008, 121, 157–168. [Google Scholar] [CrossRef]
- Abdullah, A.S.; Moffat, C.S.; Lopez-Ruiz, F.J.; Gibberd, M.R.; Hamblin, J.; Zerihun, A. Host-multi-pathogen warfare: Pathogen interactions in co-infected plants. Front. Plant Sci. 2017, 8, 1806. [Google Scholar] [CrossRef]
- Rantsiou, K.; Mataragas, M.; Alessandria, V.; Cocolin, L. Expression of virulence genes of Listeria monocytogenes in food. J. Food Saf. 2012, 32, 161–168. [Google Scholar] [CrossRef]
- Rantsiou, K.; Greppi, A.; Garosi, M.; Acquadro, A.; Mataragas, M.; Cocolin, L. Strain dependent expression of stress response and virulence genes of Listeria monocytogenes in meat juices as determined by microarray. Int. J. Food Microbiol. 2012, 152, 116–122. [Google Scholar] [CrossRef]
- Alessandria, V.; Rantsiou, K.; Dolci, P.; Zeppa, G.; Cocolin, L. Comparison of gene expression of Listeria monocytogenes in vitro and in the soft cheese Crescenza. Int. J. Dairy Technol. 2013, 66, 83–89. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Molfeta, C.; Panagiotopoulou, O.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Expression of Listeria monocytogenes key virulence genes during growth in liquid medium, on rocket and melon at 4, 10 and 30 °C. Food Microbiol. 2016, 55, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Hadjilouka, A.; Nikolidakis, K.; Paramithiotis, S.; Drosinos, E.H. Effect of co-culture with enterocinogenic E. faecium on L. monocytogenes key virulence gene expression. AIMS Microbiol. 2016, 2, 304–315. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Mavrogiannis, G.; Mallouchos, A.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Effect of lemongrass essential oil on Listeria monocytogenes gene expression. LWT 2017, 77, 510–516. [Google Scholar] [CrossRef]
- Cleveland, T.E.; Yu, J.; Fedorova, N.; Bhatnagar, D.; Payne, G.A.; Nierman, W.C.; Bennett, J.W. Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 2009, 27, 151–157. [Google Scholar] [CrossRef]
- Yu, J.; Fedorova, N.D.; Montalbano, B.G.; Bhatnagar, D.; Cleveland, T.E.; Bennett, J.W.; Nierman, W.C. Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol. Lett. 2011, 322, 145–149. [Google Scholar] [CrossRef]
- Georgiadou, M.; Dimou, A.; Yanniotis, S. Aflatoxin contamination in pistachio nuts: A farm to storage study. Food Control 2012, 26, 580–586. [Google Scholar] [CrossRef]
- Lappa, I.K.; Mparampouti, S.; Lanza, B.; Panagou, E.Z. Control of Aspergillus carbonarius in grape berries by Lactobacillus plantarum: A phenotypic and gene transcription study. Int. J. Food Microbiol. 2018, 275, 56–65. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]
- Pfaffl, M.W. Quantification strategies in real-time PCR. In A–Z of Quantitative PCR; Bustin, S.A., Ed.; International University Line (IUL): La Jolla, CA, USA, 2004; pp. 89–113. [Google Scholar]
- Kubista, M.; Sindelka, R.; Tichopad, A.; Bergkvist, A.; Lindh, D.; Forootan, A. The prime technique. real-time PCR data analysis. GIT Lab J. Eur. 2007, 11, 33–35. [Google Scholar]
- Braga, R.M.; Dourado, M.N.; Araújo, W.L. Microbial interactions: Ecology in a molecular perspective. Braz. J. Microbiol. 2016, 47 (Suppl. 1), 86–98. [Google Scholar] [CrossRef]
- Barrett, L.G.; Kniskern, J.M.; Bodenhausen, N.; Zhang, W.; Bergelson, J. Continua of specificity and virulence in plant host-pathogen interactions: Causes and consequences. New Phytol. 2009, 183, 513–529. [Google Scholar] [CrossRef] [PubMed]
- Ipcho, S.; Sundelin, T.; Erbs, G.; Kistler, H.C.; Newman, M.-A.; Olsson, S. Fungal innate immunity induced by bacterial microbe-associated molecular patterns (MAMPs). G3 2016, 6, 1585–1595. [Google Scholar] [CrossRef] [PubMed]
- Astoreca, A.; Vaamonde, G.; Dalcero, A.; Marin, S.; Ramos, A. Abiotic factors and their interactions influence on the co-production of aflatoxin B1 and cyclopiazonic acid by Aspergillus flavus isolated from corn. Food Microbiol. 2014, 38, 276–283. [Google Scholar] [CrossRef]
- Medina, A.; Gilbert, M.K.; Mack, B.M.; Obrian, G.R.; Rodríguez, A.; Bhatnagar, D.; Payne, G.; Magan, N. Interactions between water activity and temperature on the Aspergillus flavus transcriptome and aflatoxin B1 production. Int. J. Food Microbiol. 2017, 256, 36–44. [Google Scholar] [CrossRef]
- Lahouar, A.; Marin, S.; Crespo-Sempere, A.; Saïd, S.; Sanchis, V. Effects of temperature, water activity and incubation time on fungal growth and aflatoxin B1 production by toxinogenic Aspergillus flavus isolates on sorghum seeds. Rev. Argent. Microbiol. 2016, 48, 78–85. [Google Scholar] [CrossRef]
- Aldars-García, L.; Marín, S.; Sanchis, V.; Magan, N.; Medina, A. Assessment of intraspecies variability in fungal growth initiation of Aspergillus flavus and aflatoxin B1 production under static and changing temperature levels using different initial conidial inoculum levels. Int. J. Food Microbiol. 2018, 272, 1–11. [Google Scholar] [CrossRef]
- Falade, T.D.O.; Syed Mohdhamdan, S.H.; Sultanbawa, Y.; Fletcher, M.T.; Harvey, J.J.W.; Chaliha, M.; Fox, G.P. In vitro experimental environments lacking or containing soil disparately affect competition experiments of Aspergillus flavus and co-occurring fungi in maize grains. Food Addit. Contam. Part A 2016, 33, 1241–1253. [Google Scholar] [CrossRef]
- Jurado, M.; Marín, P.; Magan, N.; González-Jaén, M.T. Relationship between solute and matric potential stress, temperature, growth, and gene expression in two Fusarium verticillioides strains from Spain. Appl. Environ. Microbiol. 2008, 74, 2032–2036. [Google Scholar] [CrossRef]
- Schmidt-Heydt, M.; Abdel-Hadi, A.; Magan, N.; Geisen, R. Complex regulation of the aflatoxin biosynthesis gene cluster of Aspergillus flavus in relation to various combinations of water activity and temperature. Int. J. Food Microbiol. 2009, 135, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Heydt, M.; Magan, N.; Geisen, R. Stress induction of mycotoxin biosynthesis genes by abiotic factors. FEMS Microbiol. Lett. 2008, 284, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Ponsone, M.L.; Chiotta, M.L.; Palazzini, J.M.; Combina, M.; Chulze, S. Control of ochratoxin A production in grapes. Toxins 2012, 4, 364–372. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, G.; Shitut, S.; Preussger, D.; Yousif, G.; Waschina, S.; Kost, C. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 2018, 35, 455–488. [Google Scholar] [CrossRef] [PubMed]
- Asurmendi, P.; Barberis, C.; Pascual, L.; Dalcero, A.; Barberis, L. Influence of Listeria monocytogenes and environmental abiotic factors on growth parameters and aflatoxin B1 production by Aspergillus flavus. J. Stored Prod. Res. 2015, 60, 60–66. [Google Scholar] [CrossRef]
- Verheecke, C.; Liboz, T.; Anson, P.; Diaz, R.; Mathieu, F. Reduction of aflatoxin production by Aspergillus flavus and Aspergillus parasiticus in interaction with Streptomyces. Microbiology 2015, 161, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Lappa, I.K.; Kizis, D.; Panagou, E.Z. Monitoring the temporal expression of genes involved in ochratoxin A production of Aspergillus carbonarius under the influence of temperature and water activity. Toxins 2017, 9, 296. [Google Scholar] [CrossRef]
- Papa, K.E. Norsolorinic acid mutant of Aspergillus flavus. Microbiology 1982, 128, 1345–1348. [Google Scholar] [CrossRef]
- Bhatnagar, D.; Ehrlich, K.C.; Cleveland, T.E. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2003, 61, 83–93. [Google Scholar] [CrossRef]
- Georgianna, D.R.; Payne, G.A. Genetic regulation of aflatoxin biosynthesis: From gene to genome. Fungal Genet. Biol. 2009, 46, 113–125. [Google Scholar] [CrossRef]
- Peromingo, B.; Rodríguez, M.; Delgado, J.; Andrade, M.J.; Rodríguez, A. Gene expression as a good indicator of aflatoxin contamination in dry-cured ham. Food Microbiol. 2017, 67, 31–40. [Google Scholar] [CrossRef]
- Abdel-Hadi, A.; Schmidt-Heydt, M.; Parra, R.; Geisen, R.; Magan, N. A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by Aspergillus flavus. J. R. Soc. Interface 2012, 9, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Magan, N.; Aldred, D. Post-harvest control strategies: Minimizing mycotoxins in the food chain. Int. J. Food Microbiol. 2007, 119, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Benoit, I.; van den Esker, M.H.; Patyshakuliyeva, A.; Mattern, D.J.; Blei, F.; Zhou, M.; Dijksterhuis, J.; Brakhage, A.A.; Kuipers, O.P.; de Vries, R.P.; et al. Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ. Microbiol. 2015, 17, 2099–2113. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, M.; Mylonakis, E. Fungal-bacterial interactions and their relevance in health. Cell. Microbiol. 2015, 17, 1442–1446. [Google Scholar] [CrossRef]
- Domann, E.; Leimeister-Wachter, M.; Goebel, W.; Chakraborty, T. Molecular cloning, sequencing, and identification of a metalloprotease gene from Listeria monocytogenes that is species specific and physically linked to the listeriolysin gene. Infect. Immun. 1991, 59, 65–72. [Google Scholar]
- Behari, J.; Youngman, P. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect. Immun. 1998, 66, 3635–3642. [Google Scholar]
- van der Veen, S.; Abee, T. Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl. Environ. Microbiol. 2010, 76, 7854–7860. [Google Scholar] [CrossRef]
- Chan, Y.C.; Raengpradub, S.; Boor, K.J.; Wiedmann, M. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 2007, 73, 6484–6498. [Google Scholar] [CrossRef]
- Sztajer, H.; Szafranski, S.P.; Tomasch, J.; Reck, M.; Nimtz, M.; Rohde, M.; Wagner-Döbler, I. Cross-feeding and inter kingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 2014, 8, 2256–2271. [Google Scholar] [CrossRef]
- Tomada, S.; Sonego, P.; Moretto, M.; Engelen, K.; Pertot, I.; Perazzolli, M.; Puopolo, G. Dual RNA-Seq of Lysobacter capsici AZ78—Phytophthora infestans interaction shows the implementation of attack strategies by the bacterium and unsuccessful oomycete defense responses. Environ. Microbiol. 2017, 19, 4113–4125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Damore, J.A.; Gore, J. A slowly evolving host moves first in symbiotic interactions. Evol. Int. J. Org. Evol. 2011, 65, 2391–2398. [Google Scholar] [CrossRef] [PubMed]
- Giaouris, E.; Heir, E.; Desvaux, M.; Hébraud, M.; Møretrø, T.; Langsrud, S.; Doulgeraki, A.; Nychas, G.-J.; Kačániová, M.; Czaczyk, K.; et al. Intra-and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front. Microbiol. 2015, 6, 841. [Google Scholar] [CrossRef] [PubMed]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef]
- Jung, B.; Lee, S.; Ha, J.; Park, J.C.; Han, S.S.; Hwang, I.; Lee, Y.W.; Lee, J. Development of a selective medium for the fungal pathogen Fusarium graminearum using toxoflavin produced by the bacterial pathogen Burkholderia glumae. Plant Pathol. J. 2013, 29, 446–450. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lappa, I.K.; Dionysopoulou, A.M.; Paramithiotis, S.; Georgiadou, M.; Drosinos, E.H. Dual Transcriptional Profile of Aspergillus flavus during Co-Culture with Listeria monocytogenes and Aflatoxin B1 Production: A Pathogen–Pathogen Interaction. Pathogens 2019, 8, 198. https://doi.org/10.3390/pathogens8040198
Lappa IK, Dionysopoulou AM, Paramithiotis S, Georgiadou M, Drosinos EH. Dual Transcriptional Profile of Aspergillus flavus during Co-Culture with Listeria monocytogenes and Aflatoxin B1 Production: A Pathogen–Pathogen Interaction. Pathogens. 2019; 8(4):198. https://doi.org/10.3390/pathogens8040198
Chicago/Turabian StyleLappa, Iliada K., Angeliki Maria Dionysopoulou, Spiros Paramithiotis, Maria Georgiadou, and Eleftherios H. Drosinos. 2019. "Dual Transcriptional Profile of Aspergillus flavus during Co-Culture with Listeria monocytogenes and Aflatoxin B1 Production: A Pathogen–Pathogen Interaction" Pathogens 8, no. 4: 198. https://doi.org/10.3390/pathogens8040198
APA StyleLappa, I. K., Dionysopoulou, A. M., Paramithiotis, S., Georgiadou, M., & Drosinos, E. H. (2019). Dual Transcriptional Profile of Aspergillus flavus during Co-Culture with Listeria monocytogenes and Aflatoxin B1 Production: A Pathogen–Pathogen Interaction. Pathogens, 8(4), 198. https://doi.org/10.3390/pathogens8040198






