Peptide Epitope Hot Spots of CD4 T Cell Recognition Within Influenza Hemagglutinin During the Primary Response to Infection
Abstract
:1. Introduction
2. Results
2.1. HA Epitope Dominance in the Primary Response to Influenza Infection
2.2. Distribution of Dominant and Subdominant CD4 T Cell Epitopes in the HA Protein Sequence
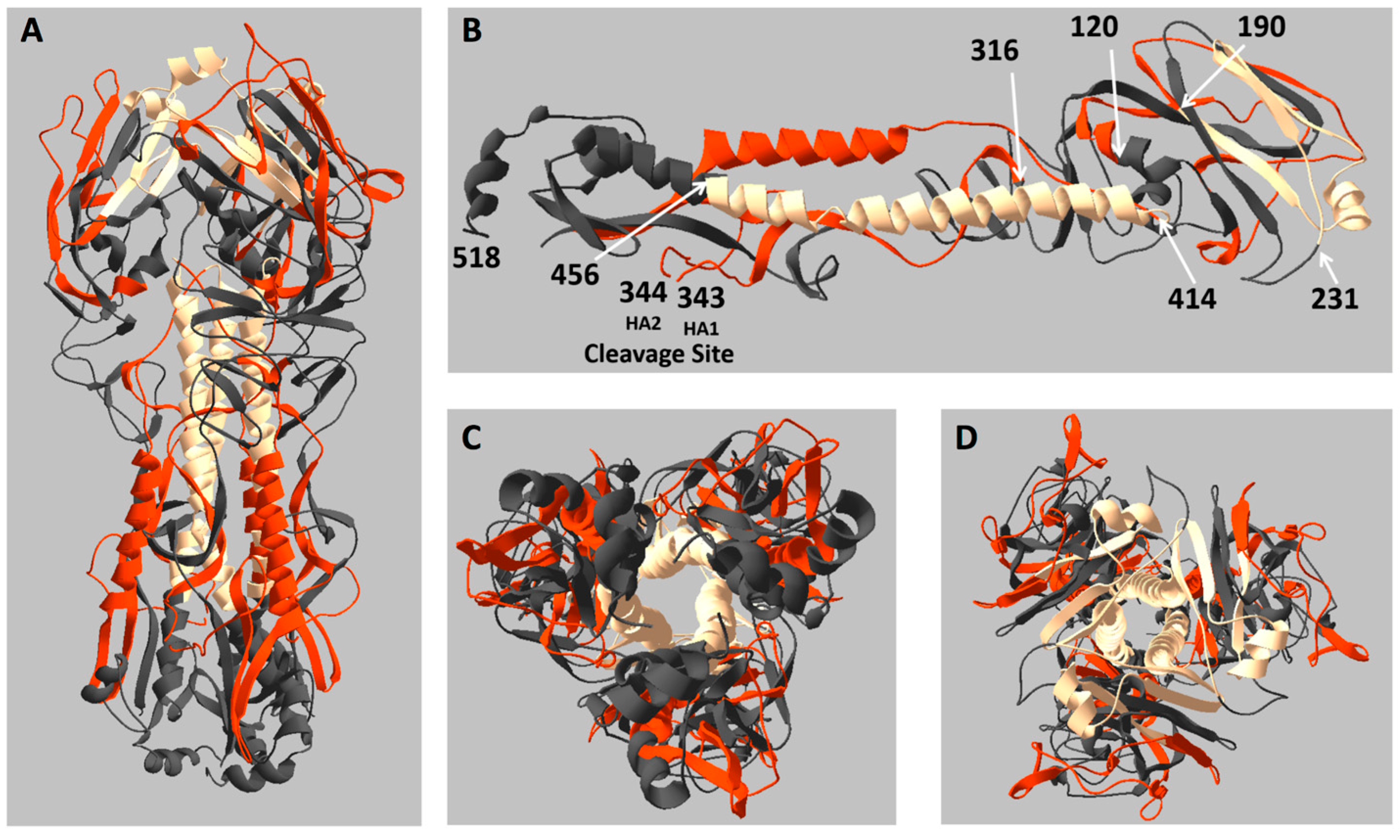
2.3. Location of Live and Dead Zones on the HA Structure
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Ethics Statement
4.3. Peptides
4.4. Infections, Tissue Harvest and Preparation of CD4 T Cell Enriched Cell Populations
4.5. EliSpot Assays
4.6. Structural Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Dunkle, L.M.; Izikson, R. Recombinant hemagglutinin influenza vaccine provides broader spectrum protection. Expert Rev. Vaccines 2016, 15, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Treanor, J.J. Clinical Practice. Influenza Vaccination. N. Engl. J. Med. 2016, 375, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Reber, A.; Katz, J. Immunological assessment of influenza vaccines and immune correlates of protection. Expert Rev. Vaccines 2013, 12, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Palese, P. Development of next generation hemagglutinin-based broadly protective influenza virus vaccines. Curr. Opin. Immunol. 2018, 53, 51–57. [Google Scholar] [CrossRef]
- Padilla-Quirarte, H.O.; Lopez-Guerrero, D.V.; Gutierrez-Xicotencatl, L.; Esquivel-Guadarrama, F. Protective Antibodies Against Influenza Proteins. Front. Immunol. 2019, 10, 1677. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xu, C.; Zhang, H.; Liu, G.D.; Xue, C.; Cao, Y. Targeting Hemagglutinin: Approaches for Broad Protection against the Influenza A Virus. Viruses 2019, 11, 405. [Google Scholar] [CrossRef] [PubMed]
- Qi, H. T follicular helper cells in space-time. Nat. Rev. Immunol. 2016, 16, 612–625. [Google Scholar] [CrossRef]
- Ueno, H. Tfh cell response in influenza vaccines in humans: What is visible and what is invisible. Curr. Opin. Immunol. 2019, 59, 9–14. [Google Scholar] [CrossRef]
- Crotty, S. A brief history of T cell help to B cells. Nat. Rev. Immunol. 2015, 15, 185–189. [Google Scholar] [CrossRef]
- Nayak, J.L.; Richards, K.A.; Yang, H.; Treanor, J.J.; Sant, A.J. Effect of influenza A(H5N1) vaccine prepandemic priming on CD4+ T-cell responses. J. Infect. Dis. 2015, 211, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Knowlden, Z.A.; Sangster, M.Y.; Sant, A.J. CD4 T cell help is limiting and selective during the primary B cell response to influenza virus infection. J. Virol. 2014, 88, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Savic, M.; Dembinski, J.L.; Kim, Y.; Tunheim, G.; Cox, R.J.; Oftung, F.; Peters, B.; Mjaaland, S. Epitope specific T-cell responses against influenza A in a healthy population. Immunology 2016, 147, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Uchtenhagen, H.; Rims, C.; Blahnik, G.; Chow, I.T.; Kwok, W.W.; Buckner, J.H.; James, E.A. Efficient ex vivo analysis of CD4+ T-cell responses using combinatorial HLA class II tetramer staining. Nat. Commun. 2016, 7, 12614. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; James, E.; Gates, T.J.; DeLong, J.H.; LaFond, R.E.; Malhotra, U.; Kwok, W.W. CD4+ T cells recognize unique and conserved 2009 H1N1 influenza hemagglutinin epitopes after natural infection and vaccination. Int. Immunol. 2013, 25, 447–457. [Google Scholar] [CrossRef]
- Roti, M.; Yang, J.; Berger, D.; Huston, L.; James, E.A.; Kwok, W.W. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J. Immunol. 2008, 180, 1758–1768. [Google Scholar] [CrossRef]
- Yang, J.; James, E.A.; Huston, L.; Danke, N.A.; Liu, A.W.; Kwok, W.W. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clin. Immunol. 2006, 120, 21–32. [Google Scholar] [CrossRef]
- Richards, K.A.; Treanor, J.J.; Nayak, J.L.; Sant, A.J. Overarching Immunodominance Patterns and Substantial Diversity in Specificity and Functionality in the Circulating Human Influenza A and B CD4 T Cell Repertoire. J. Infect. Dis. 2018. [Google Scholar] [CrossRef]
- Leddon, S.A.; Richards, K.A.; Treanor, J.J.; Sant, A.J. Abundance and specificity of influenza reactive circulating memory follicular helper and non-follicular helper CD4 T cells in healthy adults. Immunology 2015, 146, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Richards, K.A.; Nayak, J.; Chaves, F.A.; DiPiazza, A.; Knowlden, Z.A.; Alam, S.; Treanor, J.J.; Sant, A.J. Seasonal Influenza Can Poise Hosts for CD4 T-Cell Immunity to H7N9 Avian Influenza. J. Infect. Dis. 2015, 212, 86–94. [Google Scholar] [CrossRef]
- Richards, K.A.; Topham, D.; Chaves, F.A.; Sant, A.J. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J. Immunol. 2010, 185, 4998–5002. [Google Scholar] [CrossRef] [PubMed]
- Sant, A.J.; DiPiazza, A.T.; Nayak, J.L.; Rattan, A.; Richards, K.A. CD4 T cells in protection from influenza virus: Viral antigen specificity and functional potential. Immunol. Rev. 2018, 284, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Sant, A.J.; Richards, K.A.; Nayak, J. Distinct and complementary roles of CD4 T cells in protective immunity to influenza virus. Curr. Opin. Immunol. 2018, 53, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Babon, J.A.; Cruz, J.; Ennis, F.A.; Yin, L.; Terajima, M. A human CD4+ T cell epitope in the influenza hemagglutinin is cross-reactive to influenza A virus subtypes and to influenza B virus. J. Virol. 2012, 86, 9233–9243. [Google Scholar] [CrossRef] [PubMed]
- Moise, L.; Tassone, R.; Latimer, H.; Terry, F.; Levitz, L.; Haran, J.P.; Ross, T.M.; Boyle, C.M.; Martin, W.D.; De Groot, A.S. Immunization with cross-conserved H1N1 influenza CD4+ T-cell epitopes lowers viral burden in HLA DR3 transgenic mice. Hum. Vaccin. Immunother. 2013, 9, 2060–2068. [Google Scholar] [CrossRef] [Green Version]
- Alam, S.; Sant, A.J. Infection with seasonal influenza virus elicits CD4 T cells specific for genetically conserved epitopes that can be rapidly mobilized for protective immunity to pandemic H1N1 influenza virus. J. Virol. 2011, 85, 13310–13321. [Google Scholar] [CrossRef]
- Ye, M.; Kasey, S.; Khurana, S.; Nguyen, N.T.; Schubert, S.; Nugent, C.T.; Kuus-Reichel, K.; Hampl, J. MHC class II tetramers containing influenza hemagglutinin and EBV EBNA1 epitopes detect reliably specific CD4(+) T cells in healthy volunteers. Hum. Immunol. 2004, 65, 507–513. [Google Scholar] [CrossRef]
- Wang, S.F.; Yao, L.; Liu, S.J.; Chong, P.; Liu, W.T.; Chen, Y.M.; Huang, J.C. Identifying conserved DR1501-restricted CD4(+) T-cell epitopes in avian H5N1 hemagglutinin proteins. Viral. Immunol. 2010, 23, 585–593. [Google Scholar] [CrossRef]
- Duvvuri, V.R.; Moghadas, S.M.; Guo, H.; Duvvuri, B.; Heffernan, J.M.; Fisman, D.N.; Wu, G.E.; Wu, J. Highly conserved cross-reactive CD4+ T-cell HA-epitopes of seasonal and the 2009 pandemic influenza viruses. Influenza Respir. Viruses 2010, 4, 249–258. [Google Scholar] [CrossRef]
- Fehlings, M.; Chakarov, S.; Simoni, Y.; Sivasankar, B.; Ginhoux, F.; Newell, E.W. Multiplex peptide-MHC tetramer staining using mass cytometry for deep analysis of the influenza-specific T-cell response in mice. J. Immunol. Methods 2018, 453, 30–36. [Google Scholar] [CrossRef]
- Herati, R.S.; Muselman, A.; Vella, L.; Bengsch, B.; Parkhouse, K.; Del Alcazar, D.; Kotzin, J.; Doyle, S.A.; Tebas, P.; Hensley, S.E.; et al. Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucas, M.; Day, C.L.; Wyer, J.R.; Cunliffe, S.L.; Loughry, A.; McMichael, A.J.; Klenerman, P. Ex vivo phenotype and frequency of influenza virus-specific CD4 memory T cells. J. Virol. 2004, 78, 7284–7287. [Google Scholar] [CrossRef] [PubMed]
- Nayak, J.L.; Sant, A.J. Loss in CD4 T-cell responses to multiple epitopes in influenza due to expression of one additional MHC class II molecule in the host. Immunology 2012, 136, 425–436. [Google Scholar] [CrossRef]
- Nayak, J.L.; Richards, K.A.; Chaves, F.A.; Sant, A.J. Analyses of the specificity of CD4 T cells during the primary immune response to influenza virus reveals dramatic MHC-linked asymmetries in reactivity to individual viral proteins. Viral. Immunol. 2010, 23, 169–180. [Google Scholar] [CrossRef]
- Rattan, A.; Richards, K.A.; Knowlden, Z.A.G.; Sant, A.J. Protein vaccination directs the CD4+ T cell response towards shared protective epitopes that can be recalled after influenza infection. J. Virol. 2019. [Google Scholar] [CrossRef]
- Richards, K.A.; DiPiazza, A.T.; Rattan, A.; Knowlden, Z.A.G.; Yang, H.; Sant, A.J. Diverse Epitope Specificity, Immunodominance Hierarchy, and Functional Avidity of Effector CD4 T Cells Established During Priming Is Maintained in Lung After Influenza A Virus Infection. Front. Immunol. 2018, 9, 655. [Google Scholar] [CrossRef] [Green Version]
- DiPiazza, A.; Laniewski, N.; Rattan, A.; Topham, D.J.; Miller, J.; Sant, A.J. CD4 T Cell Epitope Specificity and Cytokine Potential Are Preserved as Cells Transition from the Lung Vasculature to Lung Tissue following Influenza Virus Infection. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Knowlden, Z.A.; Sant, A.J. CD4 T cell epitope specificity determines follicular versus non-follicular helper differentiation in the polyclonal response to influenza infection or vaccination. Sci. Rep. 2016, 6, 28287. [Google Scholar] [CrossRef]
- Richards, K.A.; Chaves, F.A.; Krafcik, F.R.; Topham, D.J.; Lazarski, C.A.; Sant, A.J. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J. Virol. 2007, 81, 7608–7619. [Google Scholar] [CrossRef]
- Richards, K.A.; Chaves, F.A.; Sant, A.J. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J. Virol. 2009, 83, 6566–6577. [Google Scholar] [CrossRef]
- Sundararajan, A.; Huan, L.; Richards, K.A.; Marcelin, G.; Alam, S.; Joo, H.; Yang, H.; Webby, R.J.; Topham, D.J.; Sant, A.J.; et al. Host differences in influenza-specific CD4 T cell and B cell responses are modulated by viral strain and route of immunization. PLoS ONE 2012, 7, e34377. [Google Scholar] [CrossRef] [PubMed]
- Tobery, T.W.; Wang, S.; Wang, X.M.; Neeper, M.P.; Jansen, K.U.; McClements, W.L.; Caulfield, M.J. A simple and efficient method for the monitoring of antigen-specific T cell responses using peptide pool arrays in a modified ELISpot assay. J. Immunol. Methods 2001, 254, 59–66. [Google Scholar] [CrossRef]
- Chaves, F.A.; Lee, A.H.; Nayak, J.L.; Richards, K.A.; Sant, A.J. The utility and limitations of current Web-available algorithms to predict peptides recognized by CD4 T cells in response to pathogen infection. J. Immunol. 2012, 188, 4235–4248. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Khuri, N.; Dong, G.Q.; Winter, M.B.; Shifrut, E.; Friedman, N.; Craik, C.S.; Pratt, K.P.; Paz, P.; Aswad, F.; et al. Predicting CD4 T-cell epitopes based on antigen cleavage, MHCII presentation, and TCR recognition. PLoS ONE 2018, 13, e0206654. [Google Scholar] [CrossRef] [PubMed]
- Mettu, R.R.; Charles, T.; Landry, S.J. CD4+ T-cell epitope prediction using antigen processing constraints. J. Immunol. Methods 2016, 432, 72–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, A.; Ishizuka, I.E.; Hartman, I.Z.; Poluektov, Y.; Narayan, K.; Sadegh-Nasseri, S. Studying MHC Class II Peptide Loading and Editing In Vitro. Methods Mol. Biol. 2019, 1988, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Jurewicz, M.M.; Stern, L.J. Class II MHC antigen processing in immune tolerance and inflammation. Immunogenetics 2019, 71, 171–187. [Google Scholar] [CrossRef]
- Kelly, A.; Trowsdale, J. Genetics of antigen processing and presentation. Immunogenetics 2019, 71, 161–170. [Google Scholar] [CrossRef]
- Thomas, C.; Tampe, R. Proofreading of Peptide-MHC Complexes through Dynamic Multivalent Interactions. Front. Immunol. 2017, 8, 65. [Google Scholar] [CrossRef]
- Rock, K.L.; Reits, E.; Neefjes, J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol. 2016, 37, 724–737. [Google Scholar] [CrossRef] [Green Version]
- Roche, P.A.; Cresswell, P. Antigen Processing and Presentation Mechanisms in Myeloid Cells. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Unanue, E.R.; Turk, V.; Neefjes, J. Variations in MHC Class II Antigen Processing and Presentation in Health and Disease. Annu. Rev. Immunol. 2016, 34, 265–297. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.A.; Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015, 15, 203–216. [Google Scholar] [CrossRef]
- Mintern, J.D.; Macri, C.; Villadangos, J.A. Modulation of antigen presentation by intracellular trafficking. Curr. Opin. Immunol. 2015, 34, 16–21. [Google Scholar] [CrossRef]
- Rossjohn, J.; Gras, S.; Miles, J.J.; Turner, S.J.; Godfrey, D.I.; McCluskey, J. T cell antigen receptor recognition of antigen-presenting molecules. Ann. Rev. Immunol. 2015, 33, 169–200. [Google Scholar] [CrossRef]
- Sant, A.J.; Chaves, F.A.; Jenks, S.A.; Richards, K.A.; Menges, P.; Weaver, J.M.; Lazarski, C.A. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol. Rev. 2005, 207, 261–278. [Google Scholar] [CrossRef]
- Kambayashi, T.; Laufer, T.M. Atypical MHC class II-expressing antigen-presenting cells: Can anything replace a dendritic cell? Nat. Rev. Immunol. 2014, 14, 719–730. [Google Scholar] [CrossRef]
- Kim, A.; Sadegh-Nasseri, S. Determinants of immunodominance for CD4 T cells. Curr. Opin. Immunol. 2015, 34, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Hastings, K.T. GILT: Shaping the MHC Class II-Restricted Peptidome and CD4(+) T Cell-Mediated Immunity. Front. Immunol. 2013, 4, 429. [Google Scholar] [CrossRef]
- Haque, A.; Blum, J.S. New insights in antigen processing and epitope selection: Development of novel immunotherapeutic strategies for cancer, autoimmunity and infectious diseases. J. Biol. Regul. Homeost. Agents 2005, 19, 93–104. [Google Scholar]
- Mellins, E.D.; Stern, L.J. HLA-DM and HLA-DO, key regulators of MHC-II processing and presentation. Curr. Opin. Immunol. 2014, 26, 115–122. [Google Scholar] [CrossRef]
- Pos, W.; Sethi, D.K.; Wucherpfennig, K.W. Mechanisms of peptide repertoire selection by HLA-DM. Trends Immunol 2013, 34, 495–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sant, A.J.; Chaves, F.A.; Leddon, S.A.; Tung, J. The control of the specificity of CD4 T cell responses: Thresholds, breakpoints, and ceilings. Front. Immunol. 2013, 4, 340. [Google Scholar] [CrossRef] [PubMed]
- Hassler, T.; Urmann, E.; Teschner, S.; Federle, C.; Dileepan, T.; Schober, K.; Jenkins, M.K.; Busch, D.H.; Hinterberger, M.; Klein, L. Inventories of naive and tolerant mouse CD4 T cell repertoires reveal a hierarchy of deleted and diverted T cell receptors. Proc. Natl. Acad. Sci. USA 2019, 116, 18537–18543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, M.K.; Moon, J.J. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J. Immunol 2012, 188, 4135–4140. [Google Scholar] [CrossRef] [PubMed]
- Zarnitsyna, V.I.; Evavold, B.D.; Schoettle, L.N.; Blattman, J.N.; Antia, R. Estimating the diversity, completeness, and cross-reactivity of the T cell repertoire. Front. Immunol. 2013, 4, 485. [Google Scholar] [CrossRef]
- Nielsen, M.; Lund, O.; Buus, S.; Lundegaard, C. MHC class II epitope predictive algorithms. Immunology 2010, 130, 319–328. [Google Scholar] [CrossRef]
- Vaughan, K.; Xu, X.; Caron, E.; Peters, B.; Sette, A. Deciphering the MHC-associated peptidome: A review of naturally processed ligand data. Expert Rev. Proteomics 2017, 14, 729–736. [Google Scholar] [CrossRef]
- Surman, S.; Lockey, T.D.; Slobod, K.S.; Jones, B.; Riberdy, J.M.; White, S.W.; Doherty, P.C.; Hurwitz, J.L. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc. Natl. Acad. Sci. USA 2001, 98, 4587–4592. [Google Scholar] [CrossRef]
- Koblischke, M.; Stiasny, K.; Aberle, S.W.; Malafa, S.; Tsouchnikas, G.; Schwaiger, J.; Kundi, M.; Heinz, F.X.; Aberle, J.H. Structural Influence on the Dominance of Virus-Specific CD4 T Cell Epitopes in Zika Virus Infection. Front. Immunol 2018, 9, 1196. [Google Scholar] [CrossRef]
- Avci, F.Y.; Li, X.; Tsuji, M.; Kasper, D.L. Carbohydrates and T cells: A sweet twosome. Semin. Immunol. 2013, 25, 146–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Middleton, D.R.; Wantuch, P.L.; Ozdilek, A.; Avci, F.Y. Carbohydrates as T-cell antigens with implications in health and disease. Glycobiology 2016, 26, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Kundra, R.; Kornfeld, S. Asparagine-linked oligosaccharides protect Lamp-1 and Lamp-2 from intracellular proteolysis. J. Biol. Chem. 1999, 274, 31039–31046. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.E.; Leonard, J.N. Stabilization of exosome-targeting peptides via engineered glycosylation. J. Biol. Chem. 2015, 290, 8166–8172. [Google Scholar] [CrossRef] [PubMed]
- Galli-Stampino, L.; Meinjohanns, E.; Frische, K.; Meldal, M.; Jensen, T.; Werdelin, O.; Mouritsen, S. T-cell recognition of tumor-associated carbohydrates: The nature of the glycan moiety plays a decisive role in determining glycopeptide immunogenicity. Cancer Res. 1997, 57, 3214–3222. [Google Scholar] [PubMed]
- Altman, M.O.; Angel, M.; Kosik, I.; Trovao, N.S.; Zost, S.J.; Gibbs, J.S.; Casalino, L.; Amaro, R.E.; Hensley, S.E.; Nelson, M.I.; et al. Human Influenza A Virus Hemagglutinin Glycan Evolution Follows a Temporal Pattern to a Glycan Limit. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Das, S.R.; Puigbo, P.; Hensley, S.E.; Hurt, D.E.; Bennink, J.R.; Yewdell, J.W. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog. 2010, 6, e1001211. [Google Scholar] [CrossRef]
- Tsuchiya, E.; Sugawara, K.; Hongo, S.; Matsuzaki, Y.; Muraki, Y.; Li, Z.N.; Nakamura, K. Effect of addition of new oligosaccharide chains to the globular head of influenza A/H2N2 virus haemagglutinin on the intracellular transport and biological activities of the molecule. J. Gen. Virol. 2002, 83, 1137–1146. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Ganesan, A.P.; Luckashenak, N.; Mendonca, M.; Eisenlohr, L.C. Endogenous antigen processing drives the primary CD4+ T cell response to influenza. Nat. Med. 2015, 21, 1216–1222. [Google Scholar] [CrossRef]
- Eisenlohr, L.C.; Luckashenak, N.; Apcher, S.; Miller, M.A.; Sinnathamby, G. Beyond the classical: Influenza virus and the elucidation of alternative MHC class II-restricted antigen processing pathways. Immunol. Res. 2011, 51, 237–248. [Google Scholar] [CrossRef]
- Mintern, J.D.; Villadangos, J.A. Antigen-presenting cells look within during influenza infection. Nat. Med. 2015, 21, 1123–1125. [Google Scholar] [CrossRef] [PubMed]
- Danke, N.A.; Kwok, W.W. HLA class II-restricted CD4+ T cell responses directed against influenza viral antigens postinfluenza vaccination. J. Immunol. 2003, 171, 3163–3169. [Google Scholar] [CrossRef] [PubMed]
- Howie, S.R. Blood sample volumes in child health research: Review of safe limits. Bull. World Health Organ. 2011, 89, 46–53. [Google Scholar] [CrossRef]
- Nayak, J.L.; Fitzgerald, T.F.; Richards, K.A.; Yang, H.; Treanor, J.J.; Sant, A.J. CD4+ T-cell expansion predicts neutralizing antibody responses to monovalent, inactivated 2009 pandemic influenza A(H1N1) virus subtype H1N1 vaccine. J. Infect. Dis 2013, 207, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Smart, M.; Hanson, J.; David, C.S. Characterization of HLA DR2 and DQ8 transgenic mouse with a new engineered mouse class II deletion, which lacks all endogenous class II genes. J. Autoimmun. 2003, 21, 195–199. [Google Scholar] [CrossRef]
- Stevens, J.; Corper, A.L.; Basler, C.F.; Taubenberger, J.K.; Palese, P.; Wilson, I.A. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 2004, 303, 1866–1870. [Google Scholar] [CrossRef]



| Mouse Strain | MHC Haplotype | Peptide | Amino Acids | Sequence | Spots Per 106 Cells |
|---|---|---|---|---|---|
| A/J | H-2a | HA p21 | 120–136 | 120 EQLSSVSSFERFEIFPK 136 | 474 |
| HA p30 | 174–190 | 174 YPNLSKSYVNNKEKEVL 190 | 374 | ||
| HA p37 | 215–231 | 215 VSVVSSHYSRRFTPEIA 231 | 236 | ||
| HA p56 | 328–344 | 328 LRMVTGLRNIPSIQSRG 344 | 271 | ||
| HA p61 | 358–374 | 358 TGMVDGWYGYHHQNEQG 374 | 507 | ||
| HA p64 | 375–391 | 375 SGYAADQKSTQNAINGI 391 | 351 | ||
| HA p68 | 398–414 | 398 VIEKMNTQFTAVGKEFN 414 | 640 | ||
| B10.PL | I-Au | HA p25 | 144–160 | 144 TVTGVSASCSHNGKSSF 160 | 124 |
| HA p54 | 316–332 | 316 IGECPKYVRSAKLRMVT 332 | 369 | ||
| HA p55 | 322–338 | 322 YVRSAKLRMVTGLRNIP 338 | 289 | ||
| HLA-DQ8 | DQ8 | HA p14 | 78–94 | 78 ILGNPECELLISKESWS 94 | 136 |
| HA p48 | 280–296 | 280 GIITSNAPMDECDAKCQ 296 | 155 | ||
| HLA-DR1 | DR1 | HA p27 | 156–172 | 156 GKSSFYRNLLWLTGKNG 172 | 167 |
| HA p28 | 162–178 | 162 RNLLWLTGKNGLYPNLS 178 | 163 | ||
| HA p35 | 203–219 | 203 NQRALYHTENAYVSVVS 219 | 159 | ||
| HA p74 | 434–450 | 434 IWTYNAELLVLLENERT 450 | 128 | ||
| HA p75 | 440–456 | 440 ELLVLLENERTLDFHDS 456 | 146 | ||
| BALB/c | H-2d | HA p21 | 120–136 | 120 EQLSSVSSFERFEIFPK 136 | 146 |
| HA p22 | 126–142 | 126 SSFERFEIFPKESSWPN 142 | 151 | ||
| HA p37 | 215–231 | 215 VSVVSSHYSRRFTPEIA 231 | 130 | ||
| HA p66 | 386–402 | 386 NAINGITNKVNSVIEKM 402 | 124 | ||
| SJL | H-2s | HA p21 | 120–136 | 120 EQLSSVSSFERFEIFPK 136 | 306 |
| HA p22 | 126–142 | 126 SSFERFEIFPKESSWPN 142 | 663 | ||
| HA p23 | 132–148 | 132 EIFPKESSWPNHTVTGV 148 | 300 | ||
| HA p25 | 144–160 | 144 TVTGVSASCSHNGKSSF 160 | 426 | ||
| HA p28 | 162–178 | 162 RNLLWLTGKNGLYPNLS 178 | 263 | ||
| HA p54 | 316–332 | 316 IGECPKYVRSAKLRMVT 332 | 191 | ||
| HA p57 | 334–350 | 334 LRNIPSIQSRGLFGAIA 350 | 243 | ||
| HA p66 | 386–402 | 386 NAINGITNKVNSVIEKM 402 | 428 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knowlden, Z.A.G.; Richards, K.A.; Moritzky, S.A.; Sant, A.J. Peptide Epitope Hot Spots of CD4 T Cell Recognition Within Influenza Hemagglutinin During the Primary Response to Infection. Pathogens 2019, 8, 220. https://doi.org/10.3390/pathogens8040220
Knowlden ZAG, Richards KA, Moritzky SA, Sant AJ. Peptide Epitope Hot Spots of CD4 T Cell Recognition Within Influenza Hemagglutinin During the Primary Response to Infection. Pathogens. 2019; 8(4):220. https://doi.org/10.3390/pathogens8040220
Chicago/Turabian StyleKnowlden, Zackery A. G., Katherine A. Richards, Savannah A. Moritzky, and Andrea J. Sant. 2019. "Peptide Epitope Hot Spots of CD4 T Cell Recognition Within Influenza Hemagglutinin During the Primary Response to Infection" Pathogens 8, no. 4: 220. https://doi.org/10.3390/pathogens8040220
APA StyleKnowlden, Z. A. G., Richards, K. A., Moritzky, S. A., & Sant, A. J. (2019). Peptide Epitope Hot Spots of CD4 T Cell Recognition Within Influenza Hemagglutinin During the Primary Response to Infection. Pathogens, 8(4), 220. https://doi.org/10.3390/pathogens8040220