The Role of Pseudomonas aeruginosa Lipopolysaccharide in Bacterial Pathogenesis and Physiology
Abstract
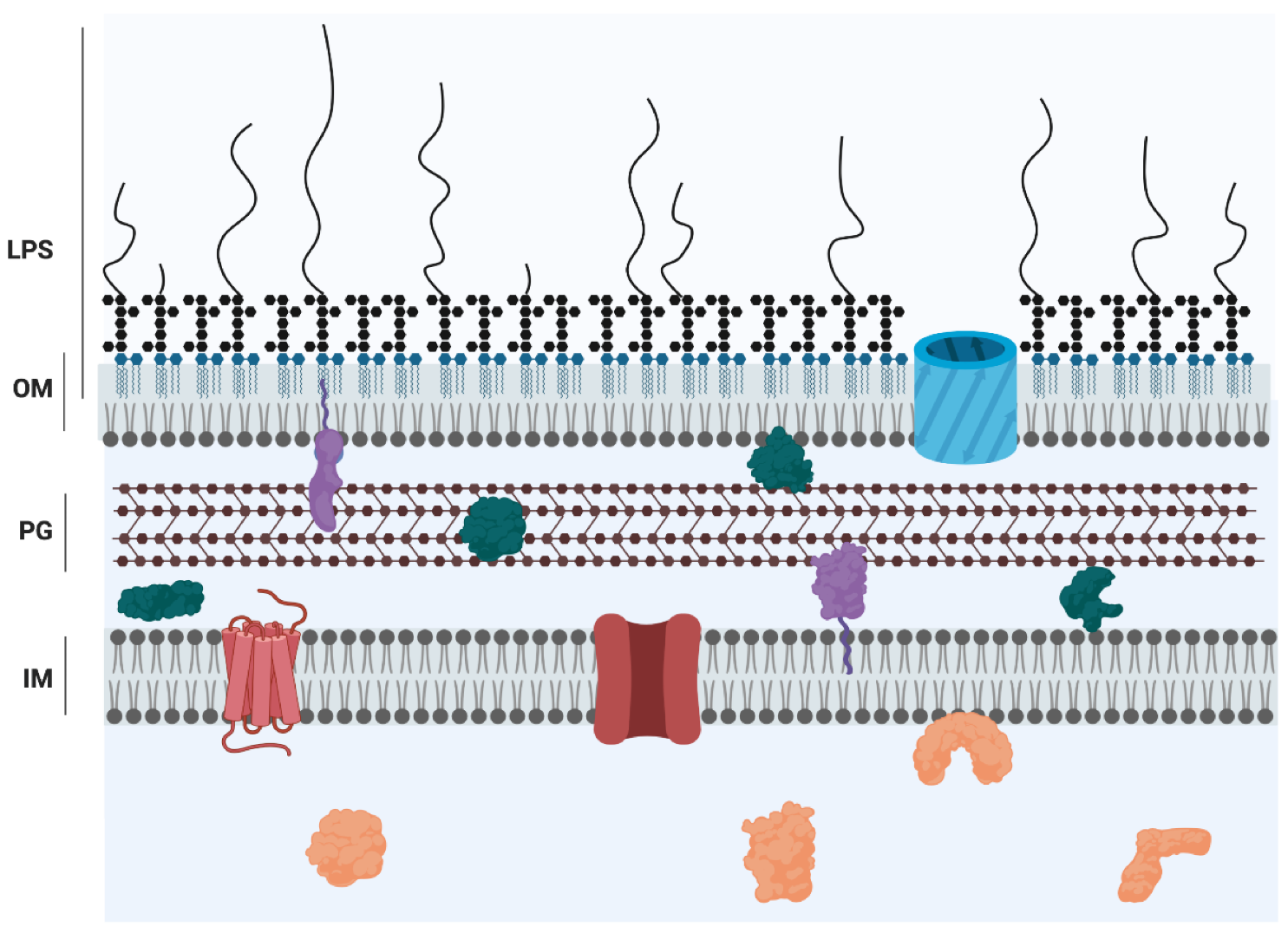
:1. Introduction
2. Structure of Lipid A, Core, and O Antigen
3. Interactions of LPS with the Host Immune System
3.1. Animal and Plant Receptors Recognize LPS and Mount an Immune Response
3.2. LPS Stimulates and Inhibits Host-Mediated Bacterial Defences
4. LPS Influences Bacterial Physiology
4.1. OMV Biogenesis and Packaging
4.2. The Role of LPS in Planktonic and Biofilm Modes of Growth
5. Antimicrobials Target LPS
5.1. Phages and Pyocins
5.2. LPS-Mediated Antibiotic Resistance
5.3. New Classes of Antibiotics Target the LPS Biosynthesis Machinery of P. aeruginosa
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Bedard, E.; Prevost, M.; Deziel, E. Pseudomonas aeruginosa in premise plumbing of large buildings. Microbiologyopen 2016, 5, 937–956. [Google Scholar] [CrossRef] [PubMed]
- Gellatly, S.L.; Hancock, R.E.W. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroth, M.N.; Cho, J.J.; Green, S.K.; Kominos, S.D.; Publishing, M.S. Epidemiology of Pseudomonas aeruginosa in agricultural areas. J. Med. Microbiol. 2018, 67, 1191–1201. [Google Scholar] [CrossRef] [PubMed]
- Cohen, T.S.; Prince, A. Cystic fibrosis: A mucosal immunodeficiency syndrome. Nat. Med. 2012, 18, 509–519. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, G.A. Cystic fibrosis airway microbiome: Overturning the old, opening the way for the new. J. Bacteriol. 2018, 200, e00561-17. [Google Scholar] [CrossRef] [Green Version]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Henderson, J.C.; Zimmerman, S.M.; Crofts, A.A.; Boll, J.M.; Kuhns, L.G.; Herrera, C.M.; Trent, M.S. The power of asymmetry: Architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu. Rev. Microbiol. 2016, 70, 255–278. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, C.; Trent, M.S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014, 83, 99–128. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.; Marx, A.; Galanos, C.; Conrad, R.S. Structural studies of lipid A from Pseudomonas aeruginosa PAO1: Occurrence of 4-amino-4-deoxyarabinose. J. Bacteriol. 1990, 172, 6631–6636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadovskaya, I.; Brisson, J.R.; Lam, J.S.; Richards, J.C.; Altman, E. Structural elucidation of the lipopolysaccharide core regions of the wild-type strain PAO1 and O-chain-deficient mutant strains AK1401 and AK1012 from Pseudomonas aeruginosa serotype O5. Eur. J. Biochem. 1998, 255, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Ernst, R.K.; Yi, E.C.; Guo, L.; Lim, K.B.; Burns, J.L.; Hackett, M.; Miller, S.I. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 1999, 286, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Kooistra, O.; Bedoux, G.; Brecker, L.; Lindner, B.; Sanchez Carballo, P.; Haras, D.; Zahringer, U. Structure of a highly phosphorylated lipopolysaccharide core in the ∆algC mutants derived from Pseudomonas aeruginosa wild-type strains PAO1 (serogroup O5) and PAC1R (serogroup O3). Carbohydr. Res. 2003, 338, 2667–2677. [Google Scholar] [CrossRef] [PubMed]
- Bystrova, O.V.; Knirel, Y.A.; Lindner, B.; Kocharova, N.A.; Kondakova, A.N.; Zähringer, U.; Pier, G.B. Structures of the core oligosaccharide and O-units in the R- and SR-type lipopolysaccharides of reference strains of Pseudomonas aeruginosa O-serogroups. FEMS Immunol. Med. Microbiol. 2006, 46, 85–99. [Google Scholar] [CrossRef] [Green Version]
- Knirel, Y.A.; Bystrova, O.V.; Kocharova, N.A.; Zähringer, U.; Pier, G.B. Review: Conserved and variable structural features in the lipopolysaccharide of Pseudomonas aeruginosa. J. Endotoxin Res. 2006, 12, 324–336. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Aebi, M.; Packer, N.H.; Seeberger, P.H.; Esko, J.D.; Stanley, P.; Hart, G.; Darvill, A.; Kinoshita, T.; et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25, 1323–1324. [Google Scholar] [CrossRef] [Green Version]
- Neelamegham, S.; Aoki-Kinoshita, K.; Bolton, E.; Frank, M.; Lisacek, F.; Lutteke, T.; O’Boyle, N.; Packer, N.H.; Stanley, P.; Toukach, P.; et al. Updates to the Symbol Nomenclature for Glycans guidelines. Glycobiology 2019, 29, 620–624. [Google Scholar] [CrossRef]
- Holst, O. Structure of the Lipopolysaccharide Core Region. In Bacterial Lipopolysaccharides: Structure, Chemical Synthesis, Biogenesis and Interaction with Host Cells; Knirel, Y.A., Valvano, M.A., Eds.; Springer Vienna: Vienna, Austria, 2011; pp. 21–39. ISBN 978-3-7091-0733-1. [Google Scholar]
- Arunmanee, W.; Pathania, M.; Solovyova, A.S.; Le Brun, A.P.; Ridley, H.; Baslé, A.; van den Berg, B.; Lakey, J.H. Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E5034–E5043. [Google Scholar] [CrossRef] [Green Version]
- Pagnout, C.; Sohm, B.; Razafitianamaharavo, A.; Caillet, C.; Offroy, M.; Leduc, M.; Gendre, H.; Jomini, S.; Beaussart, A.; Bauda, P.; et al. Pleiotropic effects of rfa-gene mutations on Escherichia coli envelope properties. Sci. Rep. 2019, 9, 9696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meredith, T.C.; Aggarwal, P.; Mamat, U.; Lindner, B.; Woodard, R.W. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Biol. 2006, 1, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Mamat, U.; Meredith, T.C.; Aggarwal, P.; Kuhl, A.; Kirchhoff, P.; Lindner, B.; Hanuszkiewicz, A.; Sun, J.; Holst, O.; Woodard, R.W. Single amino acid substitutions in either YhjD or MsbA confer viability to 3-deoxy-d-manno-oct-2-ulosonic acid-depleted Escherichia coli. Mol. Microbiol. 2008, 67, 633–648. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.M.; Raetz, C.R.H. Replacement of lipopolysaccharide with free lipid A molecules in Escherichia coli mutants lacking all core sugars. Biochemistry 2009, 48, 9627–9640. [Google Scholar] [CrossRef] [Green Version]
- Klein, G.; Lindner, B.; Brabetz, W.; Brade, H.; Raina, S. Escherichia coli K-12 Suppressor-free mutants lacking early glycosyltransferases and late acyltransferases: Minimal lipopolysaccharide structure and induction of envelope stress response. J. Biol. Chem. 2009, 284, 15369–15389. [Google Scholar] [CrossRef] [Green Version]
- Lam, J.S.; Taylor, V.L.; Islam, S.T.; Hao, Y.; Kocíncová, D. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front. Microbiol. 2011, 1, 118. [Google Scholar] [CrossRef] [Green Version]
- DebRoy, C.; Fratamico, P.M.; Yan, X.; Baranzoni, G.; Liu, Y.; Needleman, D.S.; Tebbs, R.; O’Connell, C.D.; Allred, A.; Swimley, M.; et al. Comparison of O-antigen gene clusters of all O-serogroups of Escherichia coli and proposal for adopting a new nomenclature for O-typing. PLoS ONE 2016, 11, e0147434. [Google Scholar] [CrossRef] [Green Version]
- Thrane, S.W.; Taylor, V.L.; Lund, O.; Lam, J.S.; Jelsbak, L. Application of whole-genome sequencing data for O-specific antigen analysis and in silico serotyping of Pseudomonas aeruginosa isolates. J. Clin. Microbiol. 2016, 54, 1782–1788. [Google Scholar] [CrossRef] [Green Version]
- Rivera, M.; Bryan, L.E.; Hancock, R.E.; McGroarty, E.J. Heterogeneity of lipopolysaccharides from Pseudomonas aeruginosa: Analysis of lipopolysaccharide chain length. J. Bacteriol. 1988, 170, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Yokota, S.; Kaya, S.; Sawada, S.; Kawamura, T.; Araki, Y.; Ito, E. Characterization of a polysaccharide component of lipopolysaccharide from Pseudomonas aeruginosa IID 1008 (ATCC 27584) as D-rhamnan. Eur. J. Biochem. 1987, 167, 203–209. [Google Scholar] [CrossRef]
- Weiss, J.; Barker, J. Diverse pro-inflammatory endotoxin recognition systems of mammalian innate immunity. F1000Research 2018, 7. F1000 Faculty Rev-516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathinam, V.A.K.; Zhao, Y.; Shao, F. Innate immunity to intracellular LPS. Nat. Immunol. 2019, 20, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.-S.; Lee, H.; Lee, J.-O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Cochet, F.; Peri, F. The Role of Carbohydrates in the lipopolysaccharide (LPS)/Toll-like receptor 4 (TLR4) signalling. Int. J. Mol. Sci. 2017, 18, 2318. [Google Scholar] [CrossRef] [Green Version]
- Teghanemt, A.; Zhang, D.; Levis, E.N.; Weiss, J.P.; Gioannini, T.L. Molecular basis of reduced potency of underacylated endotoxins. J. Immunol. 2005, 175, 4669–4676. [Google Scholar] [CrossRef]
- Korneev, K.V.; Arbatsky, N.P.; Molinaro, A.; Palmigiano, A.; Shaikhutdinova, R.Z.; Shneider, M.M.; Pier, G.B.; Kondakova, A.N.; Sviriaeva, E.N.; Sturiale, L.; et al. Structural relationship of the lipid A acyl groups to activation of murine Toll-like receptor 4 by lipopolysaccharides from pathogenic strains of Burkholderia mallei, Acinetobacter baumannii, and Pseudomonas aeruginosa. Front. Immunol. 2015, 6, 595. [Google Scholar] [CrossRef] [Green Version]
- Hajjar, A.M.; Ernst, R.K.; Tsai, J.H.; Wilson, C.B.; Miller, S.I. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat. Immunol. 2002, 3, 354–359. [Google Scholar] [CrossRef]
- Di Lorenzo, F.; Kubik, L.; Oblak, A.; Lore, N.I.; Cigana, C.; Lanzetta, R.; Parrilli, M.; Hamad, M.A.; De Soyza, A.; Silipo, A.; et al. Activation of human Toll-like receptor 4 (TLR4).myeloid differentiation factor 2 (MD-2) by hypoacylated lipopolysaccharide from a clinical isolate of Burkholderia cenocepacia. J. Biol. Chem. 2015, 290, 21305–21319. [Google Scholar] [CrossRef] [Green Version]
- Ernst, R.K.; Adams, K.N.; Moskowitz, S.M.; Kraig, G.M.; Kawasaki, K.; Stead, C.M.; Trent, M.S.; Miller, S.I. The Pseudomonas aeruginosa lipid A deacylase: Selection for expression and loss within the cystic fibrosis airway. J. Bacteriol. 2006, 188, 191–201. [Google Scholar] [CrossRef] [Green Version]
- Ernst, R.K.; Hajjar, A.M.; Tsai, J.H.; Moskowitz, S.M.; Wilson, C.B.; Miller, S.I. Pseudomonas aeruginosa lipid A diversity and its recognition by Toll-like receptor 4. J. Endotoxin Res. 2003, 9, 395–400. [Google Scholar] [CrossRef]
- SenGupta, S.; Hittle, L.E.; Ernst, R.K.; Uriarte, S.M.; Mitchell, T.C. A Pseudomonas aeruginosa hepta-acylated lipid A variant associated with cystic fibrosis selectively activates human neutrophils. J. Leukoc. Biol. 2016, 100, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, T.H.; Lee, M.M.; Yacono, P.W.; Cannon, C.L.; Gerceker, A.A.; Golan, D.E.; Pier, G.B. CFTR is a pattern recognition molecule that extracts Pseudomonas aeruginosa LPS from the outer membrane into epithelial cells and activates NF-kappa B translocation. Proc. Natl. Acad. Sci. USA 2002, 99, 6907–6912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pier, G.B.; Grout, M.; Zaidi, T.S.; Olsen, J.C.; Johnson, L.G.; Yankaskas, J.R.; Goldberg, J.B. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science 1996, 271, 64–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pier, G.B.; Grout, M.; Zaidi, T.S.; Goldberg, J.B. How mutant CFTR may contribute to Pseudomonas aeruginosa infection in cystic fibrosis. Am. J. Respir. Crit. Care Med. 1996, 154, S175–S182. [Google Scholar] [CrossRef] [PubMed]
- Pier, G.B.; Grout, M.; Zaidi, T.S. Cystic fibrosis transmembrane conductance regulator is an epithelial cell receptor for clearance of Pseudomonas aeruginosa from the lung. Proc. Natl. Acad. Sci. USA 1997, 94, 12088–12093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser-Pitt, D.; O’Neil, D. Cystic fibrosis-a multiorgan protein misfolding disease. Future Sci. OA 2015, 1. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, T.S.; Lyczak, J.; Preston, M.; Pier, G.B. Cystic fibrosis transmembrane conductance regulator-mediated corneal epithelial cell ingestion of Pseudomonas aeruginosa is a key component in the pathogenesis of experimental murine keratitis. Infect. Immun. 1999, 67, 1481–1492. [Google Scholar]
- Zaidi, T.; Mowrey-McKee, M.; Pier, G.B. Hypoxia increases corneal cell expression of CFTR leading to increased Pseudomonas aeruginosa binding, internalization, and initiation of inflammation. Investig. Ophthalmol. Vis. Sci. 2004, 45, 4066–4074. [Google Scholar] [CrossRef] [Green Version]
- Zaidi, T.; Bajmoczi, M.; Zaidi, T.; Golan, D.E.; Pier, G.B. Disruption of CFTR-dependent lipid rafts reduces bacterial levels and corneal disease in a murine model of Pseudomonas aeruginosa keratitis. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1000–1009. [Google Scholar] [CrossRef] [Green Version]
- Lyczak, J.B.; Zaidi, T.S.; Grout, M.; Bittner, M.; Contreras, I.; Pier, G.B. Epithelial cell contact-induced alterations in Salmonella enterica serovar Typhi lipopolysaccharide are critical for bacterial internalization. Cell. Microbiol. 2001, 3, 763–772. [Google Scholar] [CrossRef]
- Zeidler, D.; Zahringer, U.; Gerber, I.; Dubery, I.; Hartung, T.; Bors, W.; Hutzler, P.; Durner, J. Innate immunity in Arabidopsis thaliana: Lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. USA 2004, 101, 15811–15816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranf, S.; Gisch, N.; Schaffer, M.; Illig, T.; Westphal, L.; Knirel, Y.A.; Sanchez-Carballo, P.M.; Zahringer, U.; Huckelhoven, R.; Lee, J.; et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 2015, 16, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Shang-Guan, K.; Wang, M.; Htwe, N.M.P.S.; Li, P.; Li, Y.; Qi, F.; Zhang, D.; Cao, M.; Kim, C.; Weng, H.; et al. Lipopolysaccharides trigger two successive bursts of reactive oxygen species at distinct cellular locations. Plant Physiol. 2018, 176, 2543–2556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutschera, A.; Dawid, C.; Gisch, N.; Schmid, C.; Raasch, L.; Gerster, T.; Schaffer, M.; Smakowska-Luzan, E.; Belkhadir, Y.; Vlot, A.C.; et al. Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 2019, 364, 178–181. [Google Scholar] [CrossRef]
- Desaki, Y.; Kouzai, Y.; Ninomiya, Y.; Iwase, R.; Shimizu, Y.; Seko, K.; Molinaro, A.; Minami, E.; Shibuya, N.; Kaku, H.; et al. OsCERK1 plays a crucial role in the lipopolysaccharide-induced immune response of rice. New Phytol. 2018, 217, 1042–1049. [Google Scholar] [CrossRef] [Green Version]
- Iizasa, S.; Iizasa, E.; Matsuzaki, S.; Tanaka, H.; Kodama, Y.; Watanabe, K.; Nagano, Y. Arabidopsis LBP/BPI related-1 and -2 bind to LPS directly and regulate PR1 expression. Sci. Rep. 2016, 6, 27527. [Google Scholar] [CrossRef]
- Iizasa, S.; Iizasa, E.; Watanabe, K.; Nagano, Y. Transcriptome analysis reveals key roles of AtLBR-2 in LPS-induced defense responses in plants. BMC Genom. 2017, 18, 995. [Google Scholar] [CrossRef] [Green Version]
- Bajic, G.; Degn, S.E.; Thiel, S.; Andersen, G.R. Complement activation, regulation, and molecular basis for complement-related diseases. EMBO J. 2015, 34, 2735–2757. [Google Scholar] [CrossRef] [Green Version]
- Loos, M.; Bitter-Suermann, D.; Dierich, M. Interaction of the first (C1), the second (C2) and the fourth (C4) component of complement with different preparations of bacterial lipopolysaccharides and with lipid A. J. Immunol. 1974, 112, 935–940. [Google Scholar]
- Grossman, N.; Leive, L. Complement activation via the alternative pathway by purified Salmonella lipopolysaccharide is affected by its structure but not its O-antigen length. J. Immunol. 1984, 132, 376–385. [Google Scholar]
- Burns, A.M.; Hull, S.I. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect. Immun. 1998, 66, 4244–4253. [Google Scholar] [PubMed]
- Collins, M.S.; Ladehoff, D.K.; Mehton, N.S.; Noonan, J.S. Opsonic and protective activity of five human IgM monoclonal antibodies reactive with lipopolysaccharide antigen of Pseudomonas aeruginosa. FEMS Microbiol. Immunol. 1990, 2, 263–268. [Google Scholar] [CrossRef]
- Schreiber, J.R.; Cooper, L.J.; Diehn, S.; Dahlhauser, P.A.; Tosi, M.F.; Glass, D.D.; Patawaran, M.; Greenspan, N.S. Variable region-identical monoclonal antibodies of different IgG subclass directed to Pseudomonas aeruginosa lipopolysaccharide O-specific side chain function differently. J. Infect. Dis. 1993, 167, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Man-Kupisinska, A.; Swierzko, A.S.; Maciejewska, A.; Hoc, M.; Rozalski, A.; Siwinska, M.; Lugowski, C.; Cedzynski, M.; Lukasiewicz, J. Interaction of mannose-binding lectin with lipopolysaccharide outer core region and its biological consequences. Front. Immunol. 2018, 9, 1498. [Google Scholar] [CrossRef]
- Kasperkiewicz, K.; Swierzko, A.S.; Bartlomiejczyk, M.A.; Cedzynski, M.; Noszczynska, M.; Duda, K.A.; Michalski, M.; Skurnik, M. Interaction of human mannose-binding lectin (MBL) with Yersinia enterocolitica lipopolysaccharide. Int. J. Med. Microbiol. 2015, 305, 544–552. [Google Scholar] [CrossRef]
- Hong, M.; Payne, S.M. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol. Microbiol. 1997, 24, 779–791. [Google Scholar] [CrossRef]
- Bravo, D.; Silva, C.; Carter, J.A.; Hoare, A.; Alvarez, S.A.; Blondel, C.J.; Zaldívar, M.; Valvano, M.A.; Contreras, I. Growth-phase regulation of lipopolysaccharide O-antigen chain length influences serum resistance in serovars of Salmonella. J. Med. Microbiol. 2008, 57, 938–946. [Google Scholar] [CrossRef] [Green Version]
- Murray, G.L.; Attridge, S.R.; Morona, R. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 2006, 188, 2735–2739. [Google Scholar] [CrossRef] [Green Version]
- Murray, G.L.; Attridge, S.R.; Morona, R. Inducible serum resistance in Salmonella typhimurium is dependent on wzz(fepE)-regulated very long O antigen chains. Microbes Infect. 2005, 7, 1296–1304. [Google Scholar] [CrossRef]
- Murray, G.L.; Attridge, S.R.; Morona, R. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 2003, 47, 1395–1406. [Google Scholar] [CrossRef]
- Kintz, E.; Scarff, J.M.; DiGiandomenico, A.; Goldberg, J.B. Lipopolysaccharide O-antigen chain length regulation in Pseudomonas aeruginosa serogroup O11 strain PA103. J. Bacteriol. 2008, 190, 2709–2716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, I.E.; Kintz, E.N.; Porter, L.A.; Goldberg, J.B.; Burnham, N.A.; Camesano, T. A Relating the physical properties of Pseudomonas aeruginosa lipopolysaccharides to virulence by atomic force microscopy. J. Bacteriol. 2011, 193, 1259–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Prada, C.M.; Nikolich, M.; Vemulapalli, R.; Sriranganathan, N.; Boyle, S.M.; Schurig, G.G.; Hadfield, T.L.; Hoover, D.L. Deletion of wboA enhances activation of the lectin pathway of complement in Brucella abortus and Brucella melitensis. Infect. Immun. 2001, 69, 4407–4416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, A.; Isii, Y.; Tateda, K.; Matumoto, T.; Miyazaki, S.; Yokota, S.; Yamaguchi, K. Role of LPS length in clearance rate of bacteria from the bloodstream in mice. Microbiology 1995, 141, 2749–2756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiller, N.L.; Joiner, K.A. Interaction of complement with serum-sensitive and serum-resistant strains of Pseudomonas aeruginosa. Infect. Immun. 1986, 54, 689–694. [Google Scholar] [PubMed]
- Grossman, N.; Schmetz, M.A.; Foulds, J.; Klima, E.N.; Jimenez-Lucho, V.E.; Leive, L.L.; Joiner, K.A.; Jiminez, V. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J. Bacteriol. 1987, 169, 856–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joiner, K.A.; Grossman, N.; Schmetz, M.; Leive, L. C3 binds preferentially to long-chain lipopolysaccharide during alternative pathway activation by Salmonella montevideo. J. Immunol. 1986, 136, 710–715. [Google Scholar]
- Joiner, K.A.; Schmetz, M.A.; Goldman, R.C.; Leive, L.; Frank, M.M. Mechanism of bacterial resistance to complement-mediated killing: Inserted C5b-9 correlates with killing for Escherichia coli O111B4 varying in O-antigen capsule and O-polysaccharide coverage of lipid A core oligosaccharide. Infect. Immun. 1984, 45, 113–117. [Google Scholar]
- Jimenez-Lucho, V.E.; Joiner, K.A.; Foulds, J.; Frank, M.M.; Leive, L. C3b generation is affected by the structure of the O-antigen polysaccharide in lipopolysaccharide from salmonellae. J. Immunol. 1987, 139, 1253–1259. [Google Scholar]
- Wells, T.J.; Whitters, D.; Sevastsyanovich, Y.R.; Heath, J.N.; Pravin, J.; Goodall, M.; Browning, D.F.; O’Shea, M.K.; Cranston, A.; De Soyza, A.; et al. Increased severity of respiratory infections associated with elevated anti-LPS IgG2 which inhibits serum bactericidal killing. J. Exp. Med. 2014, 211, 1893–1904. [Google Scholar] [CrossRef]
- Pressler, T.; Pedersen, S.S.; Espersen, F.; Høiby, N.; Koch, C. IgG subclass antibodies to Pseudomonas aeruginosa in sera from patients with chronic Ps. aeruginosa infection investigated by ELISA. Clin. Exp. Immunol. 1990, 81, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Kronborg, G.; Pressler, T.; Fomsgaard, A.; Koch, C.; Høiby, N. Specific IgG2 antibodies to Pseudomonas aeruginosa lipid A and lipopolysaccharide are early markers of chronic infection in patients with cystic fibrosis. Infection 1993, 21, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Engels, W.; Endert, J.; Kamps, M.A.; van Boven, C.P. Role of lipopolysaccharide in opsonization and phagocytosis of Pseudomonas aeruginosa. Infect. Immun. 1985, 49, 182–189. [Google Scholar] [PubMed]
- March, C.; Cano, V.; Moranta, D.; Llobet, E.; Perez-Gutierrez, C.; Tomas, J.M.; Suarez, T.; Garmendia, J.; Bengoechea, J.A. Role of bacterial surface structures on the interaction of Klebsiella pneumoniae with phagocytes. PLoS ONE 2013, 8, e56847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.-J.; Lin, T.-L.; Hsu, C.-R.; Wang, J.-T. Use of a Dictyostelium model for isolation of genetic loci associated with phagocytosis and virulence in Klebsiella pneumoniae. Infect. Immun. 2011, 79, 997–1006. [Google Scholar] [CrossRef] [Green Version]
- Saldías, M.S.; Ortega, X.; Valvano, M.A. Burkholderia cenocepacia O antigen lipopolysaccharide prevents phagocytosis by macrophages and adhesion to epithelial cells. J. Med. Microbiol. 2009, 58, 1542–1548. [Google Scholar] [CrossRef] [Green Version]
- Eder, K.; Vizler, C.; Kusz, E.; Karcagi, I.; Glavinas, H.; Balogh, G.E.; Vigh, L.; Duda, E.; Gyorfy, Z. The role of lipopolysaccharide moieties in macrophage response to Escherichia coli. Biochem. Biophys. Res. Commun. 2009, 389, 46–51. [Google Scholar] [CrossRef]
- Burns, S.M.; Hull, S.I. Loss of resistance to ingestion and phagocytic killing by O(-) and K(-) mutants of a uropathogenic Escherichia coli O75:K5 strain. Infect. Immun. 1999, 67, 3757–3762. [Google Scholar]
- Williams, P.; Lambert, P.A.; Haigh, C.G.; Brown, M.R. The influence of the O and K antigens of Klebsiella aerogenes on surface hydrophobicity and susceptibility to phagocytosis and antimicrobial agents. J. Med. Microbiol. 1986, 21, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Duerr, C.U.; Zenk, S.F.; Chassin, C.; Pott, J.; Gutle, D.; Hensel, M.; Hornef, M.W. O-antigen delays lipopolysaccharide recognition and impairs antibacterial host defense in murine intestinal epithelial cells. PLoS Pathog. 2009, 5, e1000567. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Prada, C.M.; Zelazowska, E.B.; Nikolich, M.; Hadfield, T.L.; Roop, R.M., 2nd; Robertson, G.L.; Hoover, D.L. Interactions between Brucella melitensis and human phagocytes: Bacterial surface O-Polysaccharide inhibits phagocytosis, bacterial killing, and subsequent host cell apoptosis. Infect. Immun. 2003, 71, 2110–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porte, F.; Naroeni, A.; Ouahrani-Bettache, S.; Liautard, J.-P. Role of the Brucella suis lipopolysaccharide O antigen in phagosomal genesis and in inhibition of phagosome-lysosome fusion in murine macrophages. Infect. Immun. 2003, 71, 1481–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kus, J.V.; Tullis, E.; Cvitkovitch, D.G.; Burrows, L.L. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology 2004, 150, 1315–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korfhagen, T.R. Surfactant protein A (SP-A)-mediated bacterial clearance: SP-A and cystic fibrosis. Am. J. Respir. Cell Mol. Biol. 2001, 25, 668–672. [Google Scholar] [CrossRef]
- Rahman, S.; Gadjeva, M. Does NETosis contribute to the bacterial pathoadaptation in cystic fibrosis? Front. Immunol. 2014, 5, 378. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, E.; Rother, N.; Yanginlar, C.; Hilbrands, L.B.; van der Vlag, J. Neutrophils discriminate between lipopolysaccharides of different bacterial sources and selectively release neutrophil extracellular traps. Front. Immunol. 2016, 7, 484. [Google Scholar] [CrossRef] [Green Version]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [Green Version]
- Mashburn-Warren, L.; Howe, J.; Garidel, P.; Richter, W.; Steiniger, F.; Roessle, M.; Brandenburg, K.; Whiteley, M. Interaction of quorum signals with outer membrane lipids: Insights into prokaryotic membrane vesicle formation. Mol. Microbiol. 2008, 69, 491–502. [Google Scholar] [CrossRef] [Green Version]
- Schertzer, J.W.; Whiteley, M. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. MBio 2012, 3, e00297-11. [Google Scholar] [CrossRef] [Green Version]
- Florez, C.; Raab, J.E.; Cooke, A.C.; Schertzer, J.W. Membrane distribution of the pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. MBio 2017, 8, e01034-17. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Schertzer, J.W.; Yong, X. Molecular conformation affects the interaction of the Pseudomonas quinolone signal with the bacterial outer membrane. J. Biol. Chem. 2019, 294, 1089–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhenawy, W.; Bording-Jorgensen, M.; Valguarnera, E.; Haurat, M.F.; Wine, E.; Feldman, M.F. LPS remodeling triggers formation of outer membrane vesicles in Salmonella. MBio 2016, 7, e00940-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnington, K.E.; Kuehn, M.J. Outer membrane vesicle production facilitates LPS remodeling and outer membrane maintenance in Salmonella during environmental transitions. MBio 2016, 7, e01532-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadurugamuwa, J.L.; Beveridge, T.J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: A novel mechanism of enzyme secretion. J. Bacteriol. 1995, 177, 3998–4008. [Google Scholar] [CrossRef] [Green Version]
- Haurat, M.F.; Aduse-Opoku, J.; Rangarajan, M.; Dorobantu, L.; Gray, M.R.; Curtis, M.A.; Feldman, M.F. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 2011, 286, 1269–1276. [Google Scholar] [CrossRef] [Green Version]
- Murphy, K.; Park, A.J.; Hao, Y.; Brewer, D.; Lam, J.S.; Khursigara, C.M. Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2014, 196, 1306–1317. [Google Scholar] [CrossRef] [Green Version]
- Lasica, A.M.; Ksiazek, M.; Madej, M.; Potempa, J. The type IX secretion system (T9SS): Highlights and recent insights into its structure and function. Front. Cell. Infect. Microbiol. 2017, 7, 215. [Google Scholar] [CrossRef]
- O’Donoghue, E.J.; Sirisaengtaksin, N.; Browning, D.F.; Bielska, E.; Hadis, M.; Fernandez-Trillo, F.; Alderwick, L.; Jabbari, S.; Krachler, A.M. Lipopolysaccharide structure impacts the entry kinetics of bacterial outer membrane vesicles into host cells. PLoS Pathog. 2017, 13, e1006760. [Google Scholar] [CrossRef]
- Noh, J.-G.; Jeon, H.-E.; So, J.-S.; Chang, W.-S. Effects of the Bradyrhizobium japonicum waaL (rfaL) gene on hydrophobicity, motility, stress tolerance, and symbiotic relationship with soybeans. Int. J. Mol. Sci. 2015, 16, 16778–16791. [Google Scholar] [CrossRef] [Green Version]
- Chiku, K.; Tsunemi, K.; Yamamoto, M.; Ohnishi-Kameyama, M.; Yoshida, M.; Ishii, T.; Taguchi, F.; Iwaki, M.; Ichinose, Y.; Ono, H. Defects in D-rhamnosyl residue biosynthetic genes affect lipopolysaccharide structure, motility, and cell-surface hydrophobicity in Pseudomonas syringae pathovar glycinea race 4. Biosci. Biotechnol. Biochem. 2013, 77, 505–510. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, B.; Shi, C.; Shi, X. Mutation of a Salmonella serogroup-C1-specific gene abrogates O7-antigen biosynthesis and triggers NaCl-dependent motility deficiency. PLoS ONE 2014, 9, e106708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Hu, X.; Wang, N. The novel virulence-related gene nlxA in the lipopolysaccharide cluster of Xanthomonas citri ssp. citri is involved in the production of lipopolysaccharide and extracellular polysaccharide, motility, biofilm formation and stress resistance. Mol. Plant Pathol. 2012, 13, 923–934. [Google Scholar] [CrossRef]
- Post, D.M.B.; Yu, L.; Krasity, B.C.; Choudhury, B.; Mandel, M.J.; Brennan, C.A.; Ruby, E.G.; McFall-Ngai, M.J.; Gibson, B.W.; Apicella, M.A. O-antigen and core carbohydrate of Vibrio fischeri lipopolysaccharide: Composition and analysis of their role in Euprymna scolopes light organ colonization. J. Biol. Chem. 2012, 287, 8515–8530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgenstein, R.M.; Clemmer, K.M.; Rather, P.N. Loss of the WaaL O-antigen ligase prevents surface activation of the flagellar gene cascade in Proteus mirabilis. J. Bacteriol. 2010, 192, 3213–3221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemmer, K.M.; Bonomo, R.A.; Rather, P.N. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 2011, 157, 2534–2544. [Google Scholar] [CrossRef] [Green Version]
- Bowden, M.G.; Kaplan, H.B. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 1998, 30, 275–284. [Google Scholar] [CrossRef]
- Toguchi, A.; Siano, M.; Burkart, M.; Harshey, R.M. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: Critical role for lipopolysaccharide. J. Bacteriol. 2000, 182, 6308–6321. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Han, X.; Wang, S.; Meng, Q.; Zhang, Y.; Ding, C.; Yu, S. The waaL gene is involved in lipopolysaccharide synthesis and plays a role on the bacterial pathogenesis of avian pathogenic Escherichia coli. Vet. Microbiol. 2014, 172, 486–491. [Google Scholar] [CrossRef]
- Canals, R.; Jimenez, N.; Vilches, S.; Regue, M.; Merino, S.; Tomas, J.M. The UDP N-acetylgalactosamine 4-epimerase gene is essential for mesophilic Aeromonas hydrophila serotype O34 virulence. Infect. Immun. 2006, 74, 537–548. [Google Scholar] [CrossRef] [Green Version]
- Abeyrathne, P.D.; Daniels, C.; Poon, K.K.H.; Matewish, M.J.; Lam, J.S. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 2005, 187, 3002–3012. [Google Scholar] [CrossRef] [Green Version]
- Lindhout, T.; Lau, P.C.Y.; Brewer, D.; Lam, J.S. Truncation in the core oligosaccharide of lipopolysaccharide affects flagella-mediated motility in Pseudomonas aeruginosa PAO1 via modulation of cell surface attachment. Microbiology 2009, 155, 3449–3460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, P.C.Y.; Dutcher, J.R.; Beveridge, T.J.; Lam, J.S. Absolute quantitation of bacterial biofilm adhesion and viscoelasticity by microbead force spectroscopy. Biophys. J. 2009, 96, 2935–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, P.C.Y.; Lindhout, T.; Beveridge, T.J.; Dutcher, J.R.; Lam, J.S. Differential lipopolysaccharide core capping leads to quantitative and correlated modifications of mechanical and structural properties in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2009, 191, 6618–6631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Høiby, N. A short history of microbial biofilms and biofilm infections. APMIS 2017, 125, 272–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruhal, R.; Antti, H.; Rzhepishevska, O.; Boulanger, N.; Barbero, D.R.; Wai, S.N.; Uhlin, B.E.; Ramstedt, M. A multivariate approach to correlate bacterial surface properties to biofilm formation by lipopolysaccharide mutants of Pseudomonas aeruginosa. Colloids Surf. B. Biointerfaces 2015, 127, 182–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beveridge, T.J.; Makin, S.A.; Kadurugamuwa, J.L.; Li, Z. Interactions between biofilms and the environment. FEMS Microbiol. Rev. 1997, 20, 291–303. [Google Scholar] [CrossRef]
- Penterman, J.; Nguyen, D.; Anderson, E.; Staudinger, B.J.; Greenberg, E.P.; Lam, J.S.; Singh, P.K. Rapid evolution of culture-impaired bacteria during adaptation to biofilm growth. Cell Rep. 2014, 6, 293–300. [Google Scholar] [CrossRef] [Green Version]
- Lam, M.Y.C.; McGroarty, E.J.; Kropinski, A.M.; MacDonald, L.A.; Pedersen, S.S.; Hoiby, N.; Lam, J.S. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J. Clin. Microbiol. 1989, 27, 962–967. [Google Scholar]
- Hao, Y.; Murphy, K.; Lo, R.Y.; Khursigara, C.M.; Lam, J.S. Single-nucleotide polymorphisms found in the miga and wbpx glycosyltransferase genes account for the intrinsic lipopolysaccharide defects exhibited by Pseudomonas aeruginosa PA14. J. Bacteriol. 2015, 197, 2780–2791. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, R.R.; Mazon-Moya, M.J.; Moscoso, J.A.; Hao, Y.; Lam, J.S.; Bordi, C.; Mostowy, S.; Filloux, A. Cyclic-di-GMP regulates lipopolysaccharide modification and contributes to Pseudomonas aeruginosa immune evasion. Nat. Microbiol. 2017, 2, 17027. [Google Scholar] [CrossRef] [Green Version]
- Bedini, E.; De Castro, C.; Erbs, G.; Mangoni, L.; Dow, J.M.; Newman, M.-A.; Parrilli, M.; Unverzagt, C. Structure-dependent modulation of a pathogen response in plants by synthetic O-antigen polysaccharides. J. Am. Chem. Soc. 2005, 127, 2414–2416. [Google Scholar] [CrossRef] [PubMed]
- Clifford, J.C.; Rapicavoli, J.N.; Roper, M.C. A rhamnose-rich O-antigen mediates adhesion, virulence, and host colonization for the xylem-limited phytopathogen Xylella fastidiosa. Mol. Plant Microbe Interact. 2013, 26, 676–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapicavoli, J.N.; Blanco-Ulate, B.; Muszyński, A.; Figueroa-Balderas, R.; Morales-Cruz, A.; Azadi, P.; Dobruchowska, J.M.; Castro, C.; Cantu, D.; Roper, M.C. Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat. Commun. 2018, 9, 390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crouzet, M.; Claverol, S.; Lomenech, A.-M.; Le Senechal, C.; Costaglioli, P.; Barthe, C.; Garbay, B.; Bonneu, M.; Vilain, S. Pseudomonas aeruginosa cells attached to a surface display a typical proteome early as 20 minutes of incubation. PLoS ONE 2017, 12, e0180341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cross, A.R.; Goldberg, J.B. Remodeling of O Antigen in mucoid Pseudomonas aeruginosa via transcriptional repression of wzz2. MBio 2019, 10, e02914-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Chan, L.; Jarrell, K.; Milazzo, F.H. The nature of Pseudomonas aeruginosa strain PAO bacteriophage receptors. Can. J. Microbiol. 1977, 23, 653–658. [Google Scholar] [CrossRef]
- Jarrell, K.; Kropinski, A.M. Identification of the cell wall receptor for bacteriophage E79 in Pseudomonas aeruginosa strain PAO. J. Virol. 1977, 23, 461–466. [Google Scholar]
- Jarrell, K.F.; Kropinski, A.M. Pseudomonas aeruginosa bacteriophage phi PLS27-lipopolysaccharide interactions. J. Virol. 1981, 40, 411–420. [Google Scholar]
- Kuzio, J.; Kropinski, A.M. O-antigen conversion in Pseudomonas aeruginosa PAO1 by bacteriophage D3. J. Bacteriol. 1983, 155, 203–212. [Google Scholar]
- Temple, G.S.; Ayling, P.D.; Wilkinson, S.G. Isolation and characterization of a lipopolysaccharide-specific bacteriophage of Pseudomonas aeruginosa. Microbios 1986, 45, 81–91. [Google Scholar]
- Yokota, S.; Hayashi, T.; Matsumoto, H. Identification of the lipopolysaccharide core region as the receptor site for a cytotoxin-converting phage, phi CTX, of Pseudomonas aeruginosa. J. Bacteriol. 1994, 176, 5262–5269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Cui, X.; Zhang, F.; He, Y.; Li, L.; Yang, H. Genetic evidence for O-specific antigen as receptor of Pseudomonas aeruginosa phage K8 and its genomic analysis. Front. Microbiol. 2016, 7, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, W.S.; Phang, K.K.S.; Tan, A.H.-M.; Li, S.F.-Y.; Ow, D.S.-W. Small colony variants and single nucleotide variations in Pf1 region of PB1 phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 2016, 7, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Pan, X.; Cui, X.; Sun, Q.; Yang, X.; Yang, H. Characterization of Pseudomonas aeruginosa phage K5 genome and identification of its receptor related genes. J. Basic Microbiol. 2016, 56, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Olszak, T.; Shneider, M.M.; Latka, A.; Maciejewska, B.; Browning, C.; Sycheva, L.V.; Cornelissen, A.; Danis-Wlodarczyk, K.; Senchenkova, S.N.; Shashkov, A.S.; et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci. Rep. 2017, 7, 16302. [Google Scholar] [CrossRef] [Green Version]
- Taylor, V.L.; Fitzpatrick, A.D.; Islam, Z.; Maxwell, K.L. The diverse impacts of phage morons on bacterial fitness and virulence. Adv. Virus Res. 2019, 103, 1–31. [Google Scholar] [CrossRef]
- Newton, G.J.; Daniels, C.; Burrows, L.L.; Kropinski, A.M.; Clarke, A.J.; Lam, J.S. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol. Microbiol. 2001, 39, 1237–1247. [Google Scholar] [CrossRef]
- Taylor, V.L.; Udaskin, M.L.; Islam, S.T.; Lam, J.S. The D3 bacteriophage α-polymerase inhibitor (Iap) peptide disrupts O-antigen biosynthesis through mimicry of the chain length regulator Wzz in Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 4735–4741. [Google Scholar] [CrossRef] [Green Version]
- Roach, D.R.; Donovan, D.M. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 2015, 5, e1062590. [Google Scholar] [CrossRef] [Green Version]
- Michel-Briand, Y.; Baysse, C. The pyocins of Pseudomonas aeruginosa. Biochimie 2002, 84, 499–510. [Google Scholar] [CrossRef]
- Ghequire, M.G.K.; De Mot, R. LlpB represents a second subclass of lectin-like bacteriocins. Microb. Biotechnol. 2019, 12, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Ghequire, M.G.K.; Swings, T.; Michiels, J.; Buchanan, S.K.; De Mot, R. Hitting with a BAM: Selective killing by lectin-like bacteriocins. MBio 2018, 9, e02138-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCaughey, L.C.; Grinter, R.; Josts, I.; Roszak, A.W.; Waloen, K.I.; Cogdell, R.J.; Milner, J.; Evans, T.; Kelly, S.; Tucker, N.P.; et al. Lectin-like bacteriocins from Pseudomonas spp. utilise D-rhamnose containing lipopolysaccharide as a cellular receptor. PLoS Pathog. 2014, 10, e1003898. [Google Scholar] [CrossRef] [PubMed]
- McCaughey, L.C.; Josts, I.; Grinter, R.; White, P.; Byron, O.; Tucker, N.P.; Matthews, J.M.; Kleanthous, C.; Whitchurch, C.B.; Walker, D. Discovery, characterization and in vivo activity of pyocin SD2, a protein antibiotic from Pseudomonas aeruginosa. Biochem. J. 2016, 473, 2345–2358. [Google Scholar] [CrossRef] [Green Version]
- Köhler, T.; Donner, V.; van Delden, C. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J. Bacteriol. 2010, 192, 1921–1928. [Google Scholar] [CrossRef] [Green Version]
- Kocincova, D.; Lam, J.S. A deletion in the wapB promoter in many serotypes of Pseudomonas aeruginosa accounts for the lack of a terminal glucose residue in the core oligosaccharide and resistance to killing by R3-pyocin. Mol. Microbiol. 2013, 89, 464–478. [Google Scholar] [CrossRef]
- Salazar, A.J.; Sherekar, M.; Tsai, J.; Sacchettini, J.C. R pyocin tail fiber structure reveals a receptor-binding domain with a lectin fold. PLoS ONE 2019, 14, e0211432. [Google Scholar] [CrossRef]
- Van der Ley, P.; de Graaff, P.; Tommassen, J. Shielding of Escherichia coli outer membrane proteins as receptors for bacteriophages and colicins by O-antigenic chains of lipopolysaccharide. J. Bacteriol. 1986, 168, 449–451. [Google Scholar] [CrossRef] [Green Version]
- Hoa Tran, E.N.; Papadopoulos, M.; Morona, R. Relationship between O-antigen chain length and resistance to colicin E2 in Shigella flexneri. Microbiology 2014, 160, 589–601. [Google Scholar] [CrossRef] [Green Version]
- Sharp, C.; Boinett, C.; Cain, A.; Housden, N.G.; Kumar, S.; Turner, K.; Parkhill, J.; Kleanthous, C. O-antigen-dependent colicin insensitivity of uropathogenic Escherichia coli. J. Bacteriol. 2019, 201, e00545-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redero, M.; Lopez-Causape, C.; Aznar, J.; Oliver, A.; Blazquez, J.; Prieto, A.I. Susceptibility to R-pyocins of Pseudomonas aeruginosa clinical isolates from cystic fibrosis patients. J. Antimicrob. Chemother. 2018, 73, 2770–2776. [Google Scholar] [CrossRef] [PubMed]
- Scholl, D.; Martin, D.W.J. Antibacterial efficacy of R-type pyocins towards Pseudomonas aeruginosa in a murine peritonitis model. Antimicrob. Agents Chemother. 2008, 52, 1647–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.; Martin, L.; Rinaldi, A.; Rajendran, R.; Ramage, G.; Walker, D. Activity of pyocin S2 against Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2012, 56, 1599–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCaughey, L.C.; Ritchie, N.D.; Douce, G.R.; Evans, T.J.; Walker, D. Efficacy of species-specific protein antibiotics in a murine model of acute Pseudomonas aeruginosa lung infection. Sci. Rep. 2016, 6, 30201. [Google Scholar] [CrossRef] [PubMed]
- Paskevicius, S.; Starkevic, U.; Misiunas, A.; Vitkauskiene, A.; Gleba, Y.; Razanskiene, A. Plant-expressed pyocins for control of Pseudomonas aeruginosa. PLoS ONE 2017, 12, e0185782. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, N.M.; Martinez-Garcia, E.; Xavier, J.; Durham, W.M.; Kolter, R.; Kim, W.; Foster, K.R. Biofilm formation as a response to ecological competition. PLoS Biol. 2015, 13, e1002191. [Google Scholar] [CrossRef] [Green Version]
- Oluyombo, O.; Penfold, C.N.; Diggle, S.P. Competition in biofilms between cystic fibrosis isolates of Pseudomonas aeruginosa is shaped by R-pyocins. MBio 2019, 10, e01828-18. [Google Scholar] [CrossRef] [Green Version]
- Tümmler, B. Emerging therapies against infections with Pseudomonas aeruginosa. F1000Research 2019, 8. F1000 Faculty Rev-1371. [Google Scholar] [CrossRef]
- Hancock, R.E. Peptide antibiotics. Lancet 1997, 349, 418–422. [Google Scholar] [CrossRef]
- Simpson, B.W.; Trent, M.S. Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 2019, 17, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Srinivas, S.; Xu, Y.; Wei, W.; Feng, Y. Genetic and biochemical mechanisms for bacterial lipid A modifiers associated with polymyxin resistance. Trends Biochem. Sci. 2019, 44, 973–988. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, E.M.; O’Brien, J.P.; Brodbelt, J.S.; Trent, M.S. Extracellular zinc induces phosphoethanolamine addition to Pseudomonas aeruginosa lipid A via the ColRS two-component system. Mol. Microbiol. 2015, 97, 166–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-Y.; Chandler, C.E.; Leung, L.M.; McElheny, C.L.; Mettus, R.T.; Shanks, R.M.Q.; Liu, J.-H.; Goodlett, D.R.; Ernst, R.K.; Doi, Y. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-Negative ESKAPE pathogens. Antimicrob. Agents Chemother. 2017, 61, e00580-17. [Google Scholar] [CrossRef] [Green Version]
- Lo Sciuto, A.; Imperi, F. Aminoarabinosylation of lipid A is critical for the development of colistin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e01820-17. [Google Scholar] [CrossRef] [Green Version]
- Dosselmann, B.; Willmann, M.; Steglich, M.; Bunk, B.; Nubel, U.; Peter, S.; Neher, R.A. Rapid and consistent evolution of colistin resistance in extensively drug-resistant Pseudomonas aeruginosa during morbidostat culture. Antimicrob. Agents Chemother. 2017, 61, e00043-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jochumsen, N.; Marvig, R.L.; Damkiaer, S.; Jensen, R.L.; Paulander, W.; Molin, S.; Jelsbak, L.; Folkesson, A. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat. Commun. 2016, 7, 13002. [Google Scholar] [CrossRef] [Green Version]
- Han, M.-L.; Velkov, T.; Zhu, Y.; Roberts, K.D.; Le Brun, A.P.; Chow, S.H.; Gutu, A.D.; Moskowitz, S.M.; Shen, H.-H.; Li, J. Polymyxin-induced lipid A deacylation in Pseudomonas aeruginosa perturbs polymyxin penetration and confers high-level resistance. ACS Chem. Biol. 2018, 13, 121–130. [Google Scholar] [CrossRef]
- Yokota, S.-I.; Hakamada, H.; Yamamoto, S.; Sato, T.; Shiraishi, T.; Shinagawa, M.; Takahashi, S. Release of large amounts of lipopolysaccharides from Pseudomonas aeruginosa cells reduces their susceptibility to colistin. Int. J. Antimicrob. Agents 2018, 51, 888–896. [Google Scholar] [CrossRef]
- Manning, A.J.; Kuehn, M.J. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011, 11, 258. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilton, M.; Charron-Mazenod, L.; Moore, R.; Lewenza, S. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 544–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadurugamuwa, J.L.; Lam, J.S.; Beveridge, T.J. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effect. Antimicrob. Agents Chemother. 1993, 37, 715–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schurek, K.N.; Marr, A.K.; Taylor, P.K.; Wiegand, I.; Semenec, L.; Khaira, B.K.; Hancock, R.E.W. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 4213–4219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erwin, A.L. Antibacterial drug discovery targeting the lipopolysaccharide biosynthetic enzyme LpxC. Cold Spring Harb. Perspect. Med. 2016, 6, a025304. [Google Scholar] [CrossRef] [PubMed]
- Krause, K.M.; Haglund, C.M.; Hebner, C.; Serio, A.W.; Lee, G.; Nieto, V.; Cohen, F.; Kane, T.R.; Machajewski, T.D.; Hildebrandt, D.; et al. Potent LpxC inhibitors with in vitro activity against multi-drug resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e00977-19. [Google Scholar] [CrossRef] [PubMed]
- Cohen, F.; Aggen, J.B.; Andrews, L.D.; Assar, Z.; Boggs, J.; Choi, T.; Dozzo, P.; Easterday, A.N.; Haglund, C.M.; Hildebrandt, D.J.; et al. Optimization of LpxC inhibitors for antibacterial activity and cardiovascular safety. ChemMedChem 2019, 14, 1560–1572. [Google Scholar] [CrossRef] [PubMed]
- Achaogen Inc. Achaogen Plans for Near-Term Sale Using Structured Process through Chapter 11 of the U.S. Bankruptcy Code. Available online: http://investors.achaogen.com/news-releases/news-release-details/achaogen-plans-near-term-sale-using-structured-process-through (accessed on 26 November 2019).
- Srinivas, N.; Jetter, P.; Ueberbacher, B.J.; Werneburg, M.; Zerbe, K.; Steinmann, J.; Van der Meijden, B.; Bernardini, F.; Lederer, A.; Dias, R.L.A.; et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 2010, 327, 1010–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andolina, G.; Bencze, L.-C.; Zerbe, K.; Muller, M.; Steinmann, J.; Kocherla, H.; Mondal, M.; Sobek, J.; Moehle, K.; Malojcic, G.; et al. A peptidomimetic antibiotic interacts with the periplasmic domain of Lptd from Pseudomonas aeruginosa. ACS Chem. Biol. 2018, 13, 666–675. [Google Scholar] [CrossRef] [Green Version]
- Polyphor. Polyphor Announces Financial Results for the First Half 2019 and Realigns Strategy. Available online: https://www.polyphor.com/news/corporate-news-details/?newsid=1820271 (accessed on 26 November 2019).
- Romano, K.P.; Warrier, T.; Poulsen, B.E.; Nguyen, P.H.; Loftis, A.R.; Saebi, A.; Pentelute, B.L.; Hung, D.T. Mutations in pmrB Confer Cross-Resistance between the LptD inhibitor POL7080 and colistin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e00511-19. [Google Scholar] [CrossRef] [Green Version]
- Day, C.J.; Tran, E.N.; Semchenko, E.A.; Tram, G.; Hartley-Tassell, L.E.; Ng, P.S.K.; King, R.M.; Ulanovsky, R.; McAtamney, S.; Apicella, M.A.; et al. Glycan:glycan interactions: High affinity biomolecular interactions that can mediate binding of pathogenic bacteria to host cells. Proc. Natl. Acad. Sci. USA 2015, 112, E7266–E7275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belotserkovsky, I.; Brunner, K.; Pinaud, L.; Rouvinski, A.; Dellarole, M.; Baron, B.; Dubey, G.; Samassa, F.; Parsot, C.; Sansonetti, P.; et al. Glycan-glycan interaction determines Shigella tropism toward human T lymphocytes. MBio 2018, 9, e02309-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, E.N.H.; Day, C.J.; Poole, J.; Jennings, M.P.; Morona, R. Specific blood group antibodies inhibit Shigella flexneri interaction with human cells in the absence of spinoculation. Biochem. Biophys. Res. Commun. 2019. [Google Scholar] [CrossRef] [PubMed]
- Briard, J.G.; Jiang, H.; Moremen, K.W.; Macauley, M.S.; Wu, P. Cell-based glycan arrays for probing glycan-glycan binding protein interactions. Nat. Commun. 2018, 9, 880. [Google Scholar] [CrossRef]
- Geissner, A.; Reinhardt, A.; Rademacher, C.; Johannssen, T.; Monteiro, J.; Lepenies, B.; Thepaut, M.; Fieschi, F.; Mrazkova, J.; Wimmerova, M.; et al. Microbe-focused glycan array screening platform. Proc. Natl. Acad. Sci. USA 2019, 116, 1958–1967. [Google Scholar] [CrossRef] [Green Version]
- Hoggarth, A.; Weaver, A.; Pu, Q.; Huang, T.; Schettler, J.; Chen, F.; Yuan, X.; Wu, M. Mechanistic research holds promise for bacterial vaccines and phage therapies for Pseudomonas aeruginosa. Drug Des. Dev. Ther. 2019, 13, 909–924. [Google Scholar] [CrossRef] [Green Version]
- Kay, E.; Cuccui, J.; Wren, B.W. Recent advances in the production of recombinant glycoconjugate vaccines. NPJ Vaccines 2019, 4, 16. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huszczynski, S.M.; Lam, J.S.; Khursigara, C.M. The Role of Pseudomonas aeruginosa Lipopolysaccharide in Bacterial Pathogenesis and Physiology. Pathogens 2020, 9, 6. https://doi.org/10.3390/pathogens9010006
Huszczynski SM, Lam JS, Khursigara CM. The Role of Pseudomonas aeruginosa Lipopolysaccharide in Bacterial Pathogenesis and Physiology. Pathogens. 2020; 9(1):6. https://doi.org/10.3390/pathogens9010006
Chicago/Turabian StyleHuszczynski, Steven M., Joseph S. Lam, and Cezar M. Khursigara. 2020. "The Role of Pseudomonas aeruginosa Lipopolysaccharide in Bacterial Pathogenesis and Physiology" Pathogens 9, no. 1: 6. https://doi.org/10.3390/pathogens9010006
APA StyleHuszczynski, S. M., Lam, J. S., & Khursigara, C. M. (2020). The Role of Pseudomonas aeruginosa Lipopolysaccharide in Bacterial Pathogenesis and Physiology. Pathogens, 9(1), 6. https://doi.org/10.3390/pathogens9010006




