Effects of Naturally Occurring Mutations in Bovine Leukemia Virus 5′-LTR and Tax Gene on Viral Transcriptional Activity
Abstract
:1. Introduction
2. Results
2.1. Sequence Variations in BLV LTR Regions
2.2. BLV LTR Variants Are Determined by Specific Mutation Patterns
2.3. Increased Basal Transcriptional Activity of BLV LTRs Harboring SNPs in Regulatory Regions
2.4. Transcriptional Activities of BLV LTR Variants in the Presence of Tax344 Expression Plasmid
2.5. Variations in BLV Tax Amino Acid Sequences
2.6. Increased Promoter Activity after Transactivation with BLV Tax Variants
2.7. Ability of Tax Variants to Transactivate Different BLV LTR Variants
2.8. Association between BLV LTR and Tax Naturally Occurring Variants and Proviral Load
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Preparation
4.2. Amplification and Sequencing of LTR and Tax Gene
4.3. Sequence Data Analysis
4.4. Construction of Plasmids
4.4.1. PCR Amplification of LTR and tax Sequences
4.4.2. Cloning of LTR and Tax Sequences in Plasmid Vectors
4.5. Cell Culture
4.6. Luciferase Reporter Gene Assay
4.7. Statistical Analysis
4.8. Quantification of Proviral DNA by Real Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; Junglen, S.; et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch. Virol. 2019, 164, 2417–2429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, S.M.; Florins, A.; Gillet, N.A.; De Brogniez, A.; Sánchez-Alcaraz, M.T.; Boxus, M.; Boulanger, F.; Gutierrez, G.; Trono, K.; Alvarez, I.; et al. Preventive and Therapeutic Strategies for Bovine Leukemia Virus: Lessons for HTLV. Viruses 2011, 3, 1210–1248. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Kobayashi, S.; Konishi, M.; Kameyama, K.; Yamamoto, T.; Tsutsui, T. The recent prevalence of bovine leukemia virus (BLV) infection among Japanese cattle. Vet. Microbiol. 2011, 148, 84–88. [Google Scholar] [CrossRef] [PubMed]
- LaDronka, R.M.; Ainsworth, S.; Wilkins, M.J.; Norby, B.; Byrem, T.M.; Bartlett, P.C. Prevalence of Bovine Leukemia Virus Antibodies in US Dairy Cattle. Vet. Med. Int. 2018, 2018, 5831278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rola-Luszczak, M.; Pluta, A.; Olech, M.; Donnik, I.; Petropavlovskiy, M.; Gerilovych, A.; Vinogradova, I.; Choudhury, B.; Kuzmak, J. The molecular characterization of bovine leukaemia virus isolates from Eastern Europe and Siberia and its impact on phylogeny. PLoS ONE 2013, 8, e58705. [Google Scholar] [CrossRef]
- Ghysdael, J.; Bruck, C.; Kettmann, R.; Burny, A. Bovine leukemia virus. Curr. Top. Microbiol. Immunol. 1984, 112, 1–19. [Google Scholar]
- Sagata, N.; Yasunaga, T.; Ogawa, Y.; Tsuzuku-Kawamura, J.; Ikawa, Y. Bovine leukemia virus: Unique structural features of its long terminal repeats and its evolutionary relationship to human T-cell leukemia virus. Proc. Natl. Acad. Sci. USA 1984, 81, 4741–4745. [Google Scholar] [CrossRef] [Green Version]
- Kettmann, R.; Cleuter, Y.; Gregoire, D.; Burny, A. Role of the 3′ long open reading frame region of bovine leukemia virus in the maintenance of cell transformation. J. Virol. 1985, 54, 899–901. [Google Scholar] [CrossRef] [Green Version]
- Gillet, N.; Florins, A.; Boxus, M.; Burteau, C.; Nigro, A.; Vandermeers, F.; Balon, H.; Bouzar, A.B.; Defoiche, J.; Burny, A.; et al. Mechanisms of leukemogenesis induced by bovine leukemia virus: Prospects for novel anti-retroviral therapies in human. Retrovirology 2007, 4, 18. [Google Scholar] [CrossRef] [Green Version]
- Kincaid, R.P.; Burke, J.M.; Sullivan, C.S. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. USA 2012, 109, 3077–3082. [Google Scholar] [CrossRef] [Green Version]
- Rosewick, N.; Momont, M.; Durkin, K.; Takeda, H.; Caiment, F.; Cleuter, Y.; Vernin, C.; Mortreux, F.; Wattel, E.; Burny, A.; et al. Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 2306–2311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derse, D.; Casey, J.W. Two elements in the bovine leukemia virus long terminal repeat that regulate gene expression. Science 1986, 231, 1437–1440. [Google Scholar] [CrossRef] [PubMed]
- Couez, D.; Deschamps, J.; Kettmann, R.; Stephens, R.M.; Gilden, R.V.; Burny, A. Nucleotide sequence analysis of the long terminal repeat of integrated bovine leukemia provirus DNA and of adjacent viral and host sequences. J. Virol. 1984, 49, 615–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, I.; Yoshinaka, Y.; Ikawa, Y. Bovine leukemia virus trans-activator p38tax activates heterologous promoters with a common sequence known as a cAMP-responsive element or the binding site of a cellular transcription factor ATF. EMBO J. 1989, 8, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Merezak, C.; Pierreux, C.; Adam, E.; Lemaigre, F.; Rousseau, G.G.; Calomme, C.; Van Lint, C.; Christophe, D.; Kerkhofs, P.; Burny, A.; et al. Suboptimal enhancer sequences are required for efficient bovine leukemia virus propagation in vivo: Implications for viral latency. J. Virol. 2001, 75, 6977–6988. [Google Scholar] [CrossRef] [Green Version]
- Boros, I.M.; Tie, F.; Giam, C.Z. Interaction of bovine leukemia virus transactivator Tax with bZip proteins. Virology 1995, 214, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Willems, L.; Kettmann, R.; Chen, G.; Portetelle, D.; Burny, A.; Derse, D. A cyclic AMP-responsive DNA-binding protein (CREB2) is a cellular transactivator of the bovine leukemia virus long terminal repeat. J. Virol. 1992, 66, 766–772. [Google Scholar] [CrossRef] [Green Version]
- Brooks, P.A.; Nyborg, J.K.; Cockerell, G.L. Identification of an NF-kappa B binding site in the bovine leukemia virus promoter. J. Virol. 1995, 69, 6005–6009. [Google Scholar] [CrossRef] [Green Version]
- Niermann, G.L.; Buehring, G.C. Hormone regulation of bovine leukemia virus via the long terminal repeat. Virology 1997, 239, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Dekoninck, A.; Calomme, C.; Nizet, S.; de Launoit, Y.; Burny, A.; Ghysdael, J.; Van Lint, C. Identification and characterization of a PU.1/Spi-B binding site in the bovine leukemia virus long terminal repeat. Oncogene 2003, 22, 2882–2896. [Google Scholar] [CrossRef] [Green Version]
- Calomme, C.; Nguyen, T.L.; de Launoit, Y.; Kiermer, V.; Droogmans, L.; Burny, A.; Van Lint, C. Upstream stimulatory factors binding to an E box motif in the R region of the bovine leukemia virus long terminal repeat stimulates viral gene expression. J. Biol. Chem. 2002, 277, 8775–8789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiss-Toth, E.; Unk, I. A downstream regulatory element activates the bovine leukemia virus promoter. Biochem. Biophys. Res. Commun. 1994, 202, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M.; Temin, H.M. Lower mutation rate of bovine leukemia virus relative to that of spleen necrosis virus. J. Virol. 1994, 68, 494–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moratorio, G.; Fischer, S.; Bianchi, S.; Tome, L.; Rama, G.; Obal, G.; Carrion, F.; Pritsch, O.; Cristina, J. A detailed molecular analysis of complete bovine leukemia virus genomes isolated from B-cell lymphosarcomas. Vet. Res. 2013, 44, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coulston, J.; Naif, H.; Brandon, R.; Kumar, S.; Khan, S.; Daniel, R.C.; Lavin, M.F. Molecular cloning and sequencing of an Australian isolate of proviral bovine leukaemia virus DNA: Comparison with other isolates. J. Gen. Virol. 1990, 71, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Camargos, M.F.; Stancek, D.; Rocha, M.A.; Lessa, L.M.; Reis, J.K.; Leite, R.C. Partial sequencing of env gene of bovine leukaemia virus from Brazilian samples and phylogenetic analysis. J. Vet. Med. B Infect. Dis. Vet. Public Health 2002, 49, 325–331. [Google Scholar] [CrossRef]
- Camargos, M.F.; Pereda, A.; Stancek, D.; Rocha, M.A.; dos Reis, J.K.; Greiser-Wilke, I.; Leite, R.C. Molecular characterization of the env gene from Brazilian field isolates of Bovine leukemia virus. Virus Genes 2007, 34, 343–350. [Google Scholar] [CrossRef]
- Mamoun, R.Z.; Morisson, M.; Rebeyrotte, N.; Busetta, B.; Couez, D.; Kettmann, R.; Hospital, M.; Guillemain, B. Sequence variability of bovine leukemia virus env gene and its relevance to the structure and antigenicity of the glycoproteins. J. Virol. 1990, 64, 4180–4188. [Google Scholar] [CrossRef] [Green Version]
- Fechner, H.; Blankenstein, P.; Looman, A.C.; Elwert, J.; Geue, L.; Albrecht, C.; Kurg, A.; Beier, D.; Marquardt, O.; Ebner, D. Provirus variants of the bovine leukemia virus and their relation to the serological status of naturally infected cattle. Virology 1997, 237, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Balic, D.; Lojkic, I.; Periskic, M.; Bedekovic, T.; Jungic, A.; Lemo, N.; Roic, B.; Cac, Z.; Barbic, L.; Madic, J. Identification of a new genotype of bovine leukemia virus. Arch. Virol. 2012, 157, 1281–1290. [Google Scholar] [CrossRef]
- Polat, M.; Takeshima, S.N.; Hosomichi, K.; Kim, J.; Miyasaka, T.; Yamada, K.; Arainga, M.; Murakami, T.; Matsumoto, Y.; de la Barra Diaz, V.; et al. A new genotype of bovine leukemia virus in South America identified by NGS-based whole genome sequencing and molecular evolutionary genetic analysis. Retrovirology 2016, 13, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polat, M.; Moe, H.H.; Shimogiri, T.; Moe, K.K.; Takeshima, S.N.; Aida, Y. The molecular epidemiological study of bovine leukemia virus infection in Myanmar cattle. Arch. Virol. 2017, 162, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Todaka, H.; Uchiyama, J.; Sato, R.; Sogawa, K.; Sakaguchi, M.; Tsukamoto, K. A point mutation to the long terminal repeat of bovine leukemia virus related to viral productivity and transmissibility. Virology 2019, 537, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Grez, M.; Zornig, M.; Nowock, J.; Ziegler, M. A single point mutation activates the Moloney murine leukemia virus long terminal repeat in embryonal stem cells. J. Virol. 1991, 65, 4691–4698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Kang, S.H.; Heidenreich, O.; Brown, D.A.; Nerenberg, M.I. Sequence requirements of ATF2 and CREB binding to the human T-cell leukemia virus type 1 LTR R region. Virology 1996, 218, 362–371. [Google Scholar] [CrossRef] [Green Version]
- Pluta, A.; Rola-Luszczak, M.; Douville, R.N.; Kuzmak, J. Bovine leukemia virus long terminal repeat variability: Identification of single nucleotide polymorphisms in regulatory sequences. Virol. J. 2018, 15, 165. [Google Scholar] [CrossRef] [Green Version]
- Zyrianova, I.M.; Kovalchuk, S.N. Bovine leukemia virus tax gene/Tax protein polymorphism and its relation to Enzootic Bovine Leukosis. Virulence 2020, 11, 80–87. [Google Scholar] [CrossRef] [Green Version]
- Derse, D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J. Virol. 1987, 61, 2462–2471. [Google Scholar] [CrossRef] [Green Version]
- Rose, N.J.; Richardson, J.H.; Desselberger, U.; Lever, A.M. Virus inactivation in a proportion of human T-cell leukaemia virus type I-infected T-cell clones arises through naturally occurring mutations. J. Gen. Virol. 2000, 81, 97–104. [Google Scholar] [CrossRef]
- Niewiesk, S.; Daenke, S.; Parker, C.E.; Taylor, G.; Weber, J.; Nightingale, S.; Bangham, C.R. Naturally occurring variants of human T-cell leukemia virus type I Tax protein impair its recognition by cytotoxic T lymphocytes and the transactivation function of Tax. J. Virol. 1995, 69, 2649–2653. [Google Scholar] [CrossRef] [Green Version]
- Van Den Broeke, A.; Bagnis, C.; Ciesiolka, M.; Cleuter, Y.; Gelderblom, H.; Kerkhofs, P.; Griebel, P.; Mannoni, P.; Burny, A. In vivo rescue of a silent tax-deficient bovine leukemia virus from a tumor-derived ovine B-cell line by recombination with a retrovirally transduced wild-type tax gene. J. Virol. 1999, 73, 1054–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durkin, K.; Rosewick, N.; Artesi, M.; Hahaut, V.; Griebel, P.; Arsic, N.; Burny, A.; Georges, M.; Van den Broeke, A. Characterization of novel Bovine Leukemia Virus (BLV) antisense transcripts by deep sequencing reveals constitutive expression in tumors and transcriptional interaction with viral microRNAs. Retrovirology 2016, 13, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willems, L.; Kettmann, R.; Dequiedt, F.; Portetelle, D.; Voneche, V.; Cornil, I.; Kerkhofs, P.; Burny, A.; Mammerickx, M. In vivo infection of sheep by bovine leukemia virus mutants. J. Virol. 1993, 67, 4078–4085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajima, S.; Aida, Y. The region between amino acids 245 and 265 of the bovine leukemia virus (BLV) tax protein restricts transactivation not only via the BLV enhancer but also via other retrovirus enhancers. J. Virol. 2000, 74, 10939–10949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derse, D.; Caradonna, S.J.; Casey, J.W. Bovine leukemia virus long terminal repeat: A cell type-specific promoter. Science 1985, 227, 317–320. [Google Scholar] [CrossRef]
- Willems, L.; Kettmann, R.; Burny, A. The amino acid (157-197) peptide segment of bovine leukemia virus p34tax encompass a leucine-rich globally neutral activation domain. Oncogene 1991, 6, 159–163. [Google Scholar]
- Willems, L.; Grimonpont, C.; Heremans, H.; Rebeyrotte, N.; Chen, G.; Portetelle, D.; Burny, A.; Kettmann, R. Mutations in the bovine leukemia virus Tax protein can abrogate the long terminal repeat-directed transactivating activity without concomitant loss of transforming potential. Proc. Natl. Acad. Sci. USA 1992, 89, 3957–3961. [Google Scholar] [CrossRef] [Green Version]
- Sakakibara, N.; Kabeya, H.; Ohashi, K.; Sugimoto, C.; Onuma, M. Epitope mapping of bovine leukemia virus transactivator protein Tax. J. Vet. Med. Sci. 1998, 60, 599–605. [Google Scholar] [CrossRef] [Green Version]
- Bai, L.; Takeshima, S.N.; Isogai, E.; Kohara, J.; Aida, Y. Novel CD8(+) cytotoxic T cell epitopes in bovine leukemia virus with cattle. Vaccine 2015, 33, 7194–7202. [Google Scholar] [CrossRef] [Green Version]
- Tajima, S.; Takahashi, M.; Takeshima, S.N.; Konnai, S.; Yin, S.A.; Watarai, S.; Tanaka, Y.; Onuma, M.; Okada, K.; Aida, Y. A mutant form of the tax protein of bovine leukemia virus (BLV), with enhanced transactivation activity, increases expression and propagation of BLV in vitro but not in vivo. J. Virol. 2003, 77, 1894–1903. [Google Scholar] [CrossRef] [Green Version]
- Terme, J.M.; Calvignac, S.; Duc Dodon, M.; Gazzolo, L.; Jordan, A. E box motifs as mediators of proviral latency of human retroviruses. Retrovirology 2009, 6, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calomme, C.; Dekoninck, A.; Nizet, S.; Adam, E.; Nguyen, T.L.; Van Den Broeke, A.; Willems, L.; Kettmann, R.; Burny, A.; Van Lint, C. Overlapping CRE and E box motifs in the enhancer sequences of the bovine leukemia virus 5′ long terminal repeat are critical for basal and acetylation-dependent transcriptional activity of the viral promoter: Implications for viral latency. J. Virol. 2004, 78, 13848–13864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, P.A.; Cockerell, G.L.; Nyborg, J.K. Activation of BLV transcription by NF-kappa B and Tax. Virology 1998, 243, 94–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duckett, C.S.; Perkins, N.D.; Kowalik, T.F.; Schmid, R.M.; Huang, E.S.; Baldwin, A.S., Jr.; Nabel, G.J. Dimerization of NF-KB2 with RelA(p65) regulates DNA binding, transcriptional activation, and inhibition by an I kappa B-alpha (MAD-3). Mol. Cell Biol. 1993, 13, 1315–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emerson, R.O.; Thomas, J.H. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009, 5, e1000325. [Google Scholar] [CrossRef] [Green Version]
- Bossone, S.A.; Asselin, C.; Patel, A.J.; Marcu, K.B. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc. Natl. Acad. Sci. USA 1992, 89, 7452–7456. [Google Scholar] [CrossRef] [Green Version]
- Maity, G.; Haque, I.; Ghosh, A.; Dhar, G.; Gupta, V.; Sarkar, S.; Azeem, I.; McGregor, D.; Choudhary, A.; Campbell, D.R.; et al. The MAZ transcription factor is a downstream target of the oncoprotein Cyr61/CCN1 and promotes pancreatic cancer cell invasion via CRAF-ERK signaling. J. Biol. Chem. 2018, 293, 4334–4349. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, J.; Calame, K. The ZiN/POZ domain of ZF5 is required for both transcriptional activation and repression. Nucleic Acids Res. 1997, 25, 1108–1116. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Buehring, G.C. In vivo protein binding and functional analysis of cis-acting elements in the U3 region of the bovine leukemia virus long terminal repeat. J. Virol. 1998, 72, 5994–6003. [Google Scholar] [CrossRef] [Green Version]
- Sagata, N.; Yasunaga, T.; Tsuzuku-Kawamura, J.; Ohishi, K.; Ogawa, Y.; Ikawa, Y. Complete nucleotide sequence of the genome of bovine leukemia virus: Its evolutionary relationship to other retroviruses. Proc. Natl. Acad. Sci. USA 1985, 82, 677–681. [Google Scholar] [CrossRef] [Green Version]
- Inoue, E.; Matsumura, K.; Soma, N.; Hirasawa, S.; Wakimoto, M.; Arakaki, Y.; Yoshida, T.; Osawa, Y.; Okazaki, K. L233P mutation of the Tax protein strongly correlated with leukemogenicity of bovine leukemia virus. Vet. Microbiol. 2013, 167, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.P.; Al-Saleem, J.; Green, P.L. Comparative virology of HTLV-1 and HTLV-2. Retrovirology 2019, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Neto, W.K.; Da-Costa, A.C.; de Oliveira, A.C.; Martinez, V.P.; Nukui, Y.; Sabino, E.C.; Sanabani, S.S. Correlation between LTR point mutations and proviral load levels among human T cell lymphotropic virus type 1 (HTLV-1) asymptomatic carriers. Virol. J. 2011, 8, 535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merimi, M.; Klener, P.; Szynal, M.; Cleuter, Y.; Kerkhofs, P.; Burny, A.; Martiat, P.; Van den Broeke, A. Suppression of viral gene expression in bovine leukemia virus-associated B-cell malignancy: Interplay of epigenetic modifications leading to chromatin with a repressive histone code. J. Virol. 2007, 81, 5929–5939. [Google Scholar] [CrossRef] [Green Version]
- Achachi, A.; Florins, A.; Gillet, N.; Debacq, C.; Urbain, P.; Foutsop, G.M.; Vandermeers, F.; Jasik, A.; Reichert, M.; Kerkhofs, P.; et al. Valproate activates bovine leukemia virus gene expression, triggers apoptosis, and induces leukemia/lymphoma regression in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 10309–10314. [Google Scholar] [CrossRef] [Green Version]
- Gillet, N.A.; Gutierrez, G.; Rodriguez, S.M.; de Brogniez, A.; Renotte, N.; Alvarez, I.; Trono, K.; Willems, L. Massive depletion of bovine leukemia virus proviral clones located in genomic transcriptionally active sites during primary infection. PLoS Pathog. 2013, 9, e1003687. [Google Scholar] [CrossRef]
- Murakami, H.; Yamada, T.; Suzuki, M.; Nakahara, Y.; Suzuki, K.; Sentsui, H. Bovine leukemia virus integration site selection in cattle that develop leukemia. Virus Res. 2011, 156, 107–112. [Google Scholar] [CrossRef]
- Lemasson, I.; Polakowski, N.J.; Laybourn, P.J.; Nyborg, J.K. Transcription factor binding and histone modifications on the integrated proviral promoter in human T-cell leukemia virus-I-infected T-cells. J. Biol. Chem. 2002, 277, 49459–49465. [Google Scholar] [CrossRef] [Green Version]
- Easley, R.; Carpio, L.; Guendel, I.; Klase, Z.; Choi, S.; Kehn-Hall, K.; Brady, J.N.; Kashanchi, F. Human T-lymphotropic virus type 1 transcription and chromatin-remodeling complexes. J. Virol. 2010, 84, 4755–4768. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Murakami, H.; Uchiyama, J.; Suzuki, C.; Nikaido, S.; Shibuya, K.; Sato, R.; Maeda, Y.; Tomioka, M.; Takeshima, S.N.; Kato, H.; et al. Variations in the viral genome and biological properties of bovine leukemia virus wild-type strains. Virus Res. 2018, 253, 103–111. [Google Scholar] [CrossRef]
- Debacq, C.; Sanchez Alcaraz, M.T.; Mortreux, F.; Kerkhofs, P.; Kettmann, R.; Willems, L. Reduced proviral loads during primo-infection of sheep by Bovine Leukemia virus attenuated mutants. Retrovirology 2004, 1, 31. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wingender, E.; Karas, H.; Knuppel, R. TRANSFAC database as a bridge between sequence data libraries and biological function. Pac. Symp. Biocomput. 1997, 2, 477–485. [Google Scholar]
- Van den Broeke, A.; Cleuter, Y.; Chen, G.; Portetelle, D.; Mammerickx, M.; Zagury, D.; Fouchard, M.; Coulombel, L.; Kettmann, R.; Burny, A. Even transcriptionally competent proviruses are silent in bovine leukemia virus-induced sheep tumor cells. Proc. Natl. Acad. Sci. USA 1988, 85, 9263–9267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerkhofs, P.; Heremans, H.; Burny, A.; Kettmann, R.; Willems, L. In vitro and in vivo oncogenic potential of bovine leukemia virus G4 protein. J. Virol. 1998, 72, 2554–2559. [Google Scholar] [CrossRef] [Green Version]
- Florins, A.; Gillet, N.; Boxus, M.; Kerkhofs, P.; Kettmann, R.; Willems, L. Even attenuated bovine leukemia virus proviruses can be pathogenic in sheep. J. Virol. 2007, 81, 10195–10200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rola-Luszczak, M.; Finnegan, C.; Olech, M.; Choudhury, B.; Kuzmak, J. Development of an improved real time PCR for the detection of bovine leukaemia provirus nucleic acid and its use in the clarification of inconclusive serological test results. J. Virol. Methods 2013, 189, 258–264. [Google Scholar] [CrossRef]
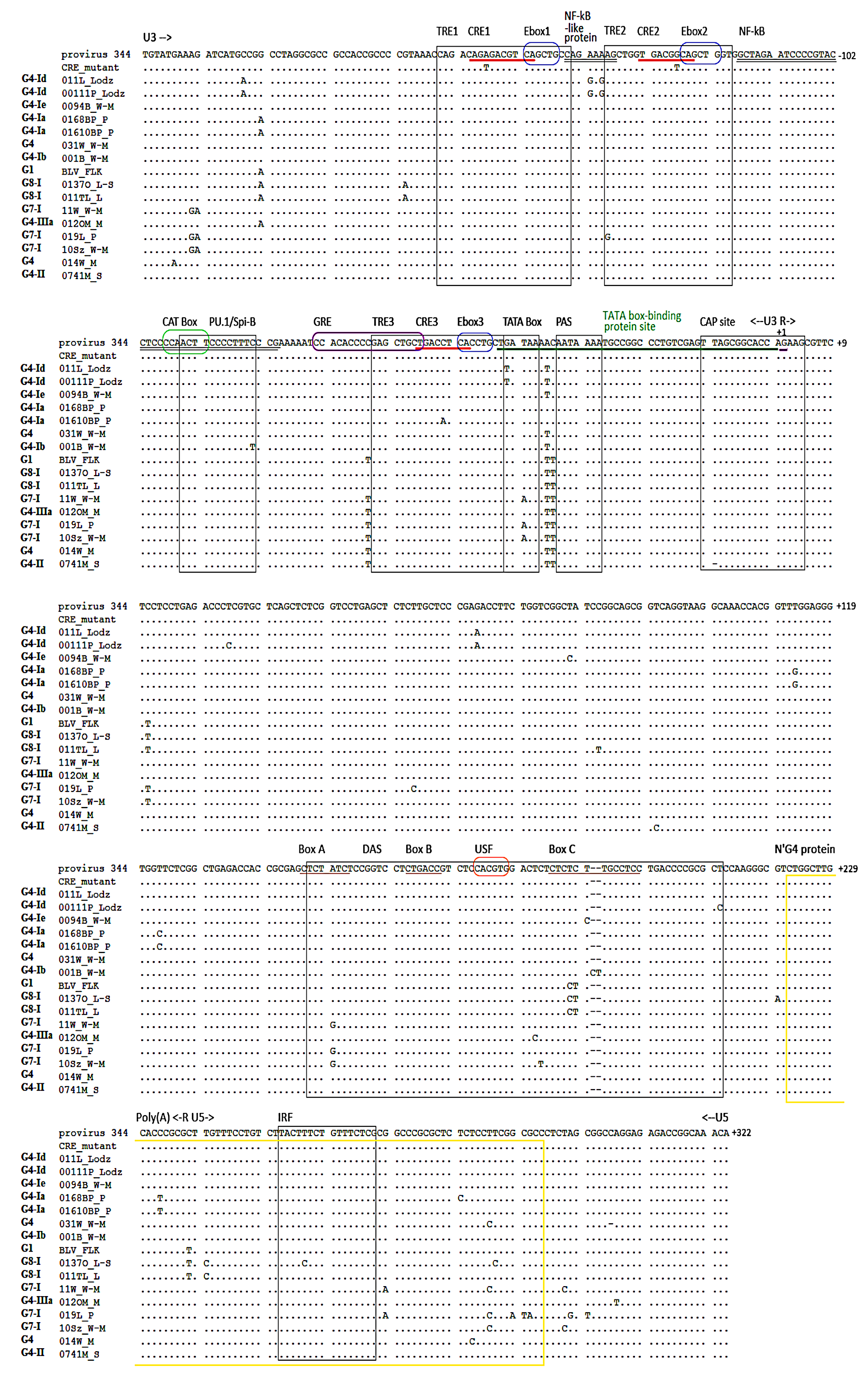









| Nucleotide Changes within LTR | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subregion | U3 | R | U5 | ||||||||||||||||||
| Regulatory Site | NF-κB-Like Protein Site | κB/ TRE2 | PU.1/ Spi-B | GRE | CRE3 | TATA Box | TATA Box-Binding Protein Site | CAP Site | Putative MAZ | DAS Box A | DAS | DAS Box C | IRF | ||||||||
| Position | −137 | −135 | −134 | −83 | −65 | −53 | −43 | −41 | −37 | −36 | −11 | +113 | +150 | +182 | +183 | +188/9 | +190 | +191/2 | +211 | +256 | |
| Sequence variants | p344 | A | A | A | C | C | C | G | T | A | C | T | T | A | T | C | TC | T | -- | T | T |
| 011L | G | G | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | -- | . | . | |
| 00111P | G | G | . | . | . | . | T | . | T | . | . | . | . | . | . | . | . | -- | C | . | |
| 0094B | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | C | -- | . | . | |
| 0168BP | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | -- | . | . | |
| 01610BP | . | . | . | . | . | A | . | . | . | . | . | G | . | . | . | . | . | -- | . | . | |
| 031W | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | -- | . | . | |
| 001B | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | . | . | CT | . | . | |
| 0137O | . | . | . | . | . | . | . | . | T | T | . | . | . | . | . | CT | . | -- | . | C | |
| 011TL | . | . | . | . | . | . | . | . | T | T | . | . | . | . | . | CT | . | -- | . | . | |
| 11W | . | . | . | . | T | . | . | A | T | T | . | . | G | . | . | . | . | -- | . | . | |
| 012OM | . | . | . | . | T | . | . | . | T | T | . | . | . | C | . | . | . | -- | . | . | |
| 019L | . | . | G | . | T | . | . | A | T | T | . | . | G | . | . | . | . | -- | . | . | |
| 10Sz | . | . | . | . | T | . | . | A | T | T | . | . | G | . | T | . | . | -- | . | . | |
| 014W | . | . | . | . | T | . | . | . | T | T | . | . | . | . | . | . | . | -- | . | . | |
| 0741M | . | . | . | . | T | . | . | . | T | T | del | . | . | . | . | . | . | -- | . | . | |
| FLK | . | . | . | . | T | . | . | . | T | T | . | . | . | . | . | . | . | -- | . | . | |
| Main/Epitope | Position | Tax Variants | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p344 | 011L | 00111P | 0094B | 0168BP | 01610BP | 031W | 001B | 0137O | 011TL | 11W | 012OM | 019L | 10Sz | 014W | 0741M | BLV- FLK | |||
| Changes within Tax | - | 56 | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | N |
| - | 64 | F | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | C | |
| - | 69 | T | M | M | M | M | M | M | . | A | A | A | . | A | A | M | . | . | |
| - | 80 | R | . | . | . | . | . | . | H | . | . | . | . | . | H | . | . | . | |
| - | 92 | P | . | . | . | . | . | . | . | . | . | . | . | . | . | . | L | . | |
| - | 93 | L | . | . | . | . | F | . | . | . | . | . | . | . | . | . | . | . | |
| - | 95 | A | . | . | . | . | . | . | . | . | . | T | . | T | T | . | . | . | |
| - | 97 | R | . | . | . | . | . | . | . | . | . | . | . | . | . | H | . | . | |
| - | 110 | Q | . | . | . | . | . | . | . | R | R | . | . | . | . | . | . | R | |
| T-cell epitope | 130 | I | . | . | . | . | . | . | . | . | . | . | . | . | . | . | L | . | |
| 140 | S | . | . | . | . | . | . | . | N | N | N | N | N | N | N | N | N | ||
| 141 | L | . | . | . | V | V | . | . | . | . | . | . | . | . | . | . | . | ||
| 142 | V | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | ||
| 148 | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | R | ||
| CTL epitope | 152 | T | . | . | . | . | . | . | . | I | I | I | I | I | I | I | I | I | |
| Leucine-rich activation domain/CTL epitopes | 164 | P | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | S | |
| 173 | L | . | . | . | . | . | . | . | . | . | P | . | P | P | . | . | . | ||
| 183 | R | . | . | . | . | . | . | . | . | . | . | . | . | . | . | K | . | ||
| 186 | I | . | . | . | . | . | . | . | T | T | . | . | . | . | . | . | . | ||
| - | 214 | I | . | . | . | . | . | . | . | . | . | . | M | . | . | . | . | . | |
| - | 221 | T | . | . | . | . | . | . | . | S | S | S | . | S | S | . | . | S | |
| - | 222 | N | . | . | . | . | . | . | . | . | . | D | . | D | D | . | . | . | |
| - | 226 | L | . | . | . | . | . | . | . | P | P | . | . | . | . | . | . | . | |
| - | 233 | L | . | . | . | . | . | F | . | I | I | P | P | P | P | . | . | . | |
| MD | 257 | C | . | . | . | . | . | . | F | . | . | G | Y | . | G | Y | Y | . | |
| 258 | D | . | . | . | . | . | . | . | . | . | . | . | N | . | . | . | . | ||
| MD/B-cell epitope | 265 | S | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | |
| - | 281 | S | . | . | . | P | P | . | . | . | . | P | . | P | P | . | . | . | |
| - | 287 | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | R | |
| - | 288 | L | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | I | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pluta, A.; Willems, L.; Douville, R.N.; Kuźmak, J. Effects of Naturally Occurring Mutations in Bovine Leukemia Virus 5′-LTR and Tax Gene on Viral Transcriptional Activity. Pathogens 2020, 9, 836. https://doi.org/10.3390/pathogens9100836
Pluta A, Willems L, Douville RN, Kuźmak J. Effects of Naturally Occurring Mutations in Bovine Leukemia Virus 5′-LTR and Tax Gene on Viral Transcriptional Activity. Pathogens. 2020; 9(10):836. https://doi.org/10.3390/pathogens9100836
Chicago/Turabian StylePluta, Aneta, Luc Willems, Renée N. Douville, and Jacek Kuźmak. 2020. "Effects of Naturally Occurring Mutations in Bovine Leukemia Virus 5′-LTR and Tax Gene on Viral Transcriptional Activity" Pathogens 9, no. 10: 836. https://doi.org/10.3390/pathogens9100836
APA StylePluta, A., Willems, L., Douville, R. N., & Kuźmak, J. (2020). Effects of Naturally Occurring Mutations in Bovine Leukemia Virus 5′-LTR and Tax Gene on Viral Transcriptional Activity. Pathogens, 9(10), 836. https://doi.org/10.3390/pathogens9100836





