Abstract
The objective of this study was to estimate the fecal carriage of Salmonella spp. among culled adult dairy cows presented to an abattoir in Wuhan, China and to evaluate their antimicrobial resistance profiles. Rectal swabs from 138 culled cows were cultured. Laboratory analysis involved the identification of Salmonella, the susceptibility assessment and the presence of Extended Spectrum β-lactamases and mcr genes in the isolates. An overall prevalence of Salmonella of 29.0% was recorded with 63.4% (26/41) and 2.4% (1/41) of the isolates identified as S. Typhimurium and S. Dublin, respectively. The occurrence of Salmonella was higher (odd ratios: 3.3) in culled cows originating from the northeast zone of China than cows originating from the central and north zones. Twenty multi-drug resistant strains (resistant to three or more antimicrobial agents) were detected (48.8%) and overall, a high resistance to ampicillin (36/41) and tetracycline (15/41) was observed. Extended Spectrum β-lactamases phenotypes were found in 7/41 isolates, of which all contained the blaCTX-M resistance gene, and no mcr genes were found by polymerase chain reaction. The high prevalence of Salmonella fecal carriage and antimicrobial resistance may contribute to an increased risk of Salmonella transmission to food.
1. Introduction
Salmonellosis is a bacterial zoonotic disease that affects a wide range of domestic animals, including chickens, pigs, and cattle, as well as humans [1,2,3]. The disease causes fever, diarrhea, dehydration, and even death among vulnerable people such as children, the elderly, and immunocompromised individuals [4,5]. In livestock, the disease manifests through various clinical signs including acute gastroenteritis (especially in young animals), abortion storms, fever, and sepsis [2,6]. In humans, Salmonella strains, other than S. Typhi and S. Paratyphi, are referred to as non-typhoidal Salmonella (NTS) and are predominately found in animal reservoirs [3,6,7]. It has been estimated that internationally, 78.7 million foodborne illness cases were caused by NTS in 2010 [8]. In dairy cattle production systems, the serovars of importance are S. Typhimurium and S. Dublin, which can be transmitted to humans via the food chain [3,6,7].
The dairy industry in China, like in most countries, is faced with increased pressures for intensification in order to meet the growing demand for dairy products and also to be able to operate profitably [9,10]. Intensification increases the risk of outbreaks of infectious diseases, such as salmonellosis, impacting negatively on both animal and human health and welfare [11]. Despite data showing Salmonella to be the second most significant bacterial foodborne pathogen in China with a prevalence of 25% in healthy dairy cattle, there is limited surveillance information on its shedding patterns, resistance profiles, and its economic impact [6,12,13]. While the use of antimicrobials plays an important role in the control and treatment of advanced Salmonella infections, the emergence of multi-drug resistance (MDR) in this bacterium poses a severe threat globally [11,14]. This resistance has been extensively documented in the poultry and pig industries [2,7,15]; however, there are few reports in cattle production systems in China [13]. Dairy products contaminated by fecal material have been implicated in sporadic cases and outbreaks in humans of non-typhoidal salmonellosis [11]. Surveillance programs involving sampling animals presented to abattoirs are not only beneficial to provide information on the frequency of carriage, animal groups at risk, and antimicrobial resistance profiles of isolates, but can also assist in aiding critical decision making as part of food safety.
Extended-spectrum β-lactamases (ESBLs) are the enzymes responsible for causing resistance to antibiotics such as penicillins and cephalosporins [16]. Globally, the important ESBL genes that have been reported to be responsible for multiple antibiotic resistance include plasmid-mediated blaCTX-M, blaTEM, blaSHV and blaKPC genes [17]. Recent studies in China have demonstrated a high prevalence of ESBLs in Escherichia coli from cattle [18,19,20,21]. There is a need to monitor the presence of ESBL in Salmonella in cattle raised under various production systems. Therefore, the objectives of this study were to estimate the occurrence (prevalence) of Salmonella in culled adult dairy cows presented to an abattoir in Wuhan, China and to evaluate their antimicrobial resistance profiles as part of a decision support tool investigating the occurrence of Salmonella in dairy farms in China. It was hypothesized that the majority of the culled cows would be positive for Salmonella.
2. Results
2.1. General Description
A total of 138 culled adult dairy cows were sampled from the 393 presented to the abattoir during the period of this study. A total of 134 animals were from north, northeast, northwest, east, and central China, with the location of 4 animals not specified.
2.2. Occurrence and Prevalence of Salmonella
The animal level prevalence of Salmonella was 29.0% (40/138; 95% confidence intervals (CI): 21.6, 37.3), with one cow having two isolates cultured (total 41 isolates). Further characterization revealed 63.4% (26/41; 95% CI: 46.9, 77.9) and 2.4% (1/41; 95% CI: 0.1, 12.9) of the Salmonella isolates to be S. Typhimurium and S. Dublin, respectively. The remaining 14 Salmonella isolates could not be characterized further. The prevalence of Salmonella in culled cows ≥5 years (32.6%; 95% CI: 23.2, 43.2) was similar to that for younger cows <5 years (21.7%; 95% CI: 10.9, 36.4) (odd ratios (OR): 1.7; 95% CI: 0.8, 4.0).
Salmonella was more commonly isolated from the culled cows sourced from the northeast zone (47.5%; 95% CI: 31.5, 63.9) compared to the central (8.5%; 95% CI: 6.3, 38.1) and north (23.1%; 95% CI: 13.5, 35.2) zones (p = 0.009) (Table 1).

Table 1.
The place of origin of the culled dairy cows sampled for the prevalence of Salmonella and multi-drug resistant (MDR) Salmonella (n = 40).
2.3. Antimicrobial Resistance Test
A detailed description of the percentage of the 41 Salmonella isolates’ resistance to nine agents is shown in Table 2. Detailed information on the antimicrobial resistance and susceptibility profiles of the 41 isolates can be found in the Supplementary Material.

Table 2.
Percentages of the resistance of 41 Salmonella spp. isolates to nine tested antibiotics.
Sensitivity towards all the tested antimicrobials was observed only in 2/41 (4.9%, 95% CI: 0.6, 16.5) of the isolates. MDR was recorded in 20 isolates (48.8%, 95% CI: 32.9, 64.9) (Table 1).
2.4. ESBL Characterization
ESBL-producing Salmonella was found in seven (17.1%; 95% CI: 7.2, 32.1) of the isolates and nine (22.0%; 95% CI: 10.6, 37.6) isolates possessed the blaCTX-M gene. No blaTEM, blaSHV, blaKPC, mcr-1, or mcr-2 were detected in any of the 41 Salmonella isolates. Eight of the nine blaCTX-M positive isolates were cultured from cows sourced from the north and northeast zones of China.
3. Discussion
Salmonella was cultured from approximately 30% of the sampled culled adult dairy cows at the abattoir in Wuhan, China. The prevalence recorded from this study was slightly higher than that reported in other studies from the USA (25%; 3%) and Italy (0%) in culled dairy cows [22,23,24]. In China, a prevalence of approximately 20% was reported in cattle sampled at multiple abattoirs and it was postulated that the feces and the hides posed a high risk for contamination with Salmonella [13]. Yang [25] found 13 Salmonella isolates from 78 pieces of beef in marketplaces in Shaanxi and Xu [26] reported the prevalence of Salmonella in beef increased recently (9.30–15.79%) in southern China. The high prevalence of Salmonella in feces in this research may be related to the place of origin.
With data collected by the National Foodborne Disease Outbreaks Surveillance System, Salmonella was reported to be responsible for 13.2% of all human foodborne cases in China in 2013 [12]. The per capita consumption of beef (predominantly sourced from beef cattle, culled dairy cows, and sometimes buffalo) is predicted to increase in China [9], with intensification of domestic production systems and international importation required to meet this demand [27]. Culled dairy cows comprise a large part of beef production in many countries. Currently, for example, in Canada and the USA, approximately 30% of beef production is sourced from culled dairy cows [28,29]. The high prevalence of Salmonella detected in the current study is a potential risk to humans, as well as increasing the risk of cross-contamination of carcasses within the abattoir environment [13]. There is a need to determine the occurrence of Salmonella within China’s dairy farms and to investigate potential risk factors so that appropriate mitigation strategies can be developed and implemented [13,30,31].
From this study, about 50% of the isolated Salmonella demonstrated multi-drug resistance. The recorded multi-drug resistance in this study was higher than an earlier study conducted with cattle in China (0% and 35%, respectively) [6,13] but was lower than that found in dry milk-related infant food (75%) [32]. The antimicrobials with the highest resistance were ampicillin and tetracycline. These antibiotics belonged to the two most commonly used antibiotic groups in 2018 in the Chinese livestock industries, including chicken and pigs [33]. This finding is similar to that reported in culled dairy cows in California [22] and is likely linked to the global long-term use of these antibiotics for promoting growth and controlling mastitis in dairy cows [34].
The findings of this study showed high antimicrobial resistance towards ampicillin (AMP) and tetracyclines (TET). From this study, polymyxin B (PB) and imipenem (IPM) were observed to have the least resistance. These findings are in agreement with which the findings of others [25,35,36]. PB had been used as a therapeutic drug and feed additive since the 1980s in China. Its usage is expected to reach 16 500 tonnes by the year 2021 [37]. IPM, one of the carbapenems, is widely used against multi-resistant Gram-negative pathogens in human medicine, especially strains producing ESBL [38]. The resistance profiles towards PB and IPM are of public health importance as the encoding genes responsible for resistance are located on the mobile genetic elements [37]. Our study showed a low occurrence of the resistance genes (ESBL and mcr) in Salmonella. The absence of the mcr gene in the tested Salmonella isolates is likely a result of the ban on the use of polymyxin for promoting the growth of livestock in China in 2016 [39]. The monitoring of resistance genes in bacterial isolates is important to provide evidence on the reduction in the use of antimicrobials [16]. There is a need to determine and assess antimicrobial usage patterns in dairy farms in China, as has been undertaken in other countries [22,40].
In this study, we only collected fecal swabs from culled dairy cows at the abattoir but did not collect samples from other sites, such as the hides, which have been shown to be heavily contaminated with Salmonella in another study in China [13]. We were unable to characterize the distribution of Salmonella isolates based on the origin source of the cows due to the small sample size. Both the lack of detailed data on farms and of information on transport duration and condition of transportation were further limitations to this study, as others have highlighted the importance of these factors on shedding/carriage of Salmonella [41,42]. Our study highlights the potential hazard to food safety arising from Salmonella carriage by culled dairy cows processed at an abattoir. There is a need to determine the occurrence and carriage of Salmonella among dairy cows in China across the different regions to identify potential risk factors for infection so that appropriate on-farm control measures can be developed.
4. Materials and Methods
4.1. Study Area and Approvals
This study was conducted in Wuhan, China in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (National Health and Medical Research Council, 2013) and the Guide for the Care and Use of Laboratory Animals (issued by the Committee of Hubei People’s Congress, Hubei, China, 2005) with the approval of the Animal Ethics Committee of Murdoch University (approval number R3201/19) and the Committee on the Ethics of Animal Experiments at Huazhong Agricultural University.
4.2. Study Design
This was an abattoir surveillance study conducted from June to July 2019 in Wuhan investigating the prevalence of Salmonella in fecal samples collected from adult culled dairy cows from Chinese dairy farms. The selected abattoir processed most of the culled dairy cows for local consumption in Wuhan. The source population included all culled dairy cows at the abattoir during the study time and these originated from the north (Inner Mongolia and Hebei Provinces), northeast (Heilongjiang, Jilin, and Liaoning Provinces), northwest (the Ningxia Hui Autonomous Region), central (Henan and Hubei Provinces), and east (Shandong Province) zones of China (Figure 1).
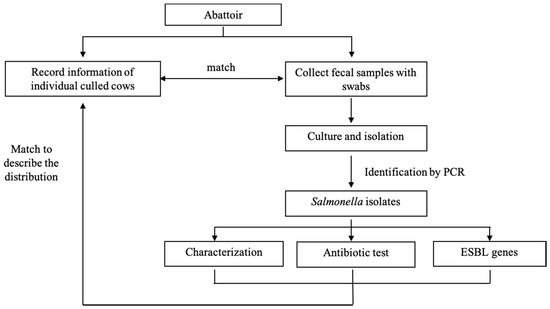
Figure 1.
Framework for the assessment of carriage of Salmonella in culled cow in abattoir.
4.3. Sample Size Calculations
Using Epitools (https://epitools.ausvet.com.au) with an assumed prevalence of 50%, 95% confidence intervals, and 10% precision a sample size of 100 animals was deemed appropriate.
4.4. Study Populations and Selection of Study Subjects
A total of 138 culled adult dairy cows were sampled in this study. All sampled cows were Holstein Friesians, the predominant dairy breed within the dairy industry in China. A total of 3–5 animals from the 6–10 animals processed each day were selected based on their originating zones. Data on the provinces of origin were recorded prior to sampling. An animal’s age was assessed by examination of their incisors, and then, categorized into four groups (2.5, 3.5, 4.5, and ≥5 years old).
4.5. Collection of Fecal Swabs
Feces were collected post stunning by inserting two cotton swabs into the rectum and rotating against the rectal wall. These were then immediately placed into a sterile tube containing 10 mL of buffered peptone water (BPW) (Hopebio Co., Ltd., Qingdao, China), stored on ice, and transported to the State Key Laboratory of Agricultural Microbiology of Huazhong Agricultural University, Wuhan, where they were processed on the day of collection.
4.6. Isolation and Identification
The culture and isolation of Salmonella on the fecal swabs were conducted according to the international standard methods: EN-ISO 6579:2002/A1: 2007: Amendment 1: Annex D. Briefly, pre-enrichment with BPW was done at 37 ℃ for 18–20 h and thereafter, 100 µl from the pre-enrichment homogenate was inserted on a modified semi-solid Rappaport–Vassiliadis (MSRV) medium (Hopebio Co., Ltd., Qingdao, China), while 1 mL of culture was transferred into 9 mL of Mueller–Kauffmann Tetrathionate novobiocin (MKTTn) broth (Hopebio Co., Ltd., Qingdao, China). Both preparations were then incubated at 41.5 and 37 ℃, respectively, for selective enrichment. After 48 h, suspected growth on the MSRV media and aliquots from all MKTTn Broth were streaked onto Xylose Lysine Deoxycholate (XLD) agar (Hopebio Co., Ltd., Qingdao, China), which was incubated aerobically at 37 °C for 24 h. At least three typical-looking colonies of Salmonella on each XLD agar plate were selected and were confirmed by performing a polymerase chain reaction (PCR) with the primers for the Salmonella enterotoxin (Stn) gene [43]. After this, Salmonella isolates with different colony morphologies but from the same sample were taken and purified for further molecular identification.
4.7. Characterization Using Rapid Molecular Detection Methods
The confirmed Salmonella were characterized using a PCR according to serotype-specific genes to detect S. Typhimurium, S. Enteritidis, S. Agona, S. Dublin, and S. Infantis [44,45,46] (Table 3).

Table 3.
Primers of the Salmonella serotype genes used in this study.
4.8. Antibiotic Susceptibility Test
Antibiotic susceptibility of the Salmonella isolates was determined by the broth microdilution method according to the guidelines recommended by the Clinical and Laboratory Standards Institute (CLSI) [47,48]. A total of 9 antimicrobials representing 9 classes that are commonly used in animals and humans were used. This included AMP, TET, GEN, IPM, and SXT with recommended cut-off values by CLSI [48] and florfenicol (FFC), enrofloxacin (ENR), and ceftiofur (CEF), which had recommended cut-off values by CLSI M31-A3 [47]. The interpreted result of PB was unavailable in the recent CLSI criteria; therefore, a PB ≥ 4 μg/mL, recognized by CLSI in 1981 [49], was considered resistant. AMP, TET, GEN, ENR, FFC, and CEF were selected as these are frequently used in veterinary clinical food animal practice. Susceptibility to PB was also tested, as it can still be used for therapeutic purposes in China, although it has been officially banned for use as a growth-promoter [21]. Escherichia coli ATCC 25,922 was used as the quality control strain. Strains resistant to three or more antimicrobial agents were considered as being MDR.
4.9. Identification of ESBL-Producing Salmonella
Each Salmonella isolate was streaked onto Mueller–Hinton Agar supplemented with 1 µg/mL cefotaxime [50]. Suspected ESBL-producing Salmonella on the Mueller–Hinton Agar were further examined by combination disk diffusion according to the CLSI guidelines using cefotaxime (30 μg) ± clavulanic acid (10 μg) and ceftazidime (30 μg) ± clavulanic acid (10 μg) [48]. All isolates were cultured in BPW for 8 h, and then, DNA was extracted from 1mL of the liquid using the boiling method and analyzed with a PCR for blaCTX-M [51], blaTEM [52], blaSHV [53], blaKPC [38], mcr-1 [37], and mcr-2 [54] genes (Table 4).

Table 4.
Primers of the ESBL- resistant genes used in this study.
4.10. Statistical Analysis
Data sorting was done in Microsoft Excel (Microsoft Office 2017) and thereafter analyzed using the statistical software SPSS version 22.0 (SPSS, Inc., Chicago, IL, USA). The prevalence of Salmonella at the animal level was estimated. The associations between age groups, locations, and patterns of resistance were determined by calculating odd ratios and their 95% CI. A Chi-square test was used to compare the prevalence between the age groups and locations and the results were interpreted at p value ≤ 0.05.
5. Conclusions
From this abattoir surveillance, it is concluded that the prevalence of Salmonella spp. among adult culled dairy cows was 29%. A high proportion (approximate 50%) of the isolates showed multiple-drug resistance. There is a need to determine the occurrence of Salmonella in dairy farms and the potential risk factors, and further develop appropriate on-farm control measures.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/10/853/s1, Table S1: Information of Salmonella isolates in the abattoir, Table S2: The combianation of multi-drug resistance.
Author Contributions
Conceptualization, I.D.R., A.G. and J.W.A.; methodology, A.G., J.W.A. and X.Z.; formal analysis, J.W., I.D.R., and J.W.A.; investigation, J.W., K.X., P.Y. and Y.P.; resources, A.G., J.W.A. and X.L.; writing—original draft preparation, J.W.; writing—review and editing, I.D.R., J.W.A. and A.G.; software, Y.C.; supervision, I.D.R., A.G. and J.W.A.; project administration, Q.P. and Z.W.; funding acquisition, J.W.A., A.G. and X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2018YFD0501605), Hubei Province Technical Innovation Special Project (Foreign Scientific and Technological Cooperation) (Grant No. 2019AHB069), the Special Fund for Chinese Agricultural Research System (beef/yaks) (CARS-37), and the National Distinguished Scholars in Agricultural Research and Technical Innovative Team. The author Jie Wang was supported by the International Education Program for Innovative Talents (2016) sponsored by the China Scholarship Council and International Tuition Fee Scholarship supplied by Murdoch University.
Acknowledgments
We thank all staff members from Murdoch University, Huazhong Agricultural University, and the local abattoir who provided technical support for this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gieraltowski, L.; Higa, J.; Peralta, V.; Green, A.; Schwensohn, C.; Rosen, H.; Libby, T.; Kissler, B.; Marsden-Haug, N.; Booth, H.; et al. Salmonella Heidelberg Investigation Team. National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS ONE 2016, 11, e0162369. [Google Scholar] [CrossRef] [PubMed]
- Jiu, Y.; Zhu, S.; Khan, S.B.; Sun, M.; Zou, G.; Meng, X.; Wu, B.; Zhou, R.; Li, S. Phenotypic and genotypic resistance of Salmonella isolates from healthy and diseased pigs in China during 2008–2015. Microb. Drug Resist. 2016, 23, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Awosile, B.; McClure, J.; Sanchez, J.; Rodriguez-Lecompte, J.C.; Keefe, G.; Heider, L.C. Salmonella enterica and extended-spectrum cephalosporin-resistant Escherichia coli recovered from holstein dairy calves from 8 farms in New Brunswick, Canada. J. Dairy. Sci. 2018, 101, 3271–3284. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Tian, L.; Cheng, Z.; Liu, W.; Li, S.; Yu, W.; Zhang, W.; Xiang, X.; Sun, Z. Viral and bacterial etiology of acute diarrhea among children under 5 years of age in Wuhan, China. Chin. Med. J. 2016, 129, 1939–1944. [Google Scholar] [CrossRef]
- Kuang, X.; Hao, H.; Dai, M.; Wang, Y.; Ahmad, I.; Liu, Z.; Yuan, Z. Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front. Microbiol. 2015, 6, 602. [Google Scholar] [CrossRef]
- Li, Y.; Pan, Z.; Kang, X.; Geng, S.; Liu, Z.; Cai, Y.; Jiao, X. Prevalence, characteristics, and antimicrobial resistance patterns of Salmonella in retail pork in Jiangsu Province, eastern China. J. Food Prot. 2014, 77, 236–245. [Google Scholar] [CrossRef]
- WHO. WHO Estimates of the Global Burden of Foodborne Disease Foodborne Diseases Burden Epidemiology Reference Group 2007–2015. Available online: http://www.who.int/foodsafety/publications/foodborne_disease/fergreport/en/ (accessed on 5 May 2017).
- National Bureau of Statistics of China (NBSC). 2018 Statistical Announcement on National Economic and Social Development. Available online: http://www.stats.gov.cn/tjsj/zxfb/201902/t20190228_1651265.html (accessed on 17 October 2020).
- Zhong, Z.; Chen, S.; Kong, X.; Tracy, M. Why improving agrifood quality is difficult in China: Evidence from dairy industry. China Econ. Rev. 2014, 31, 74–83. [Google Scholar] [CrossRef]
- Laufer, A.S.; Grass, J.; Holt, K.; Whichard, J.M.; Griffin, P.M.; Gould, L.H. Outbreaks of Salmonella infections attributed to beef—United States, 1973–2011. Epidemiol. Infect. 2015, 143, 2003–2013. [Google Scholar] [CrossRef]
- Li, W.W.; Wang, S.T.; Liang, J.J.; Liu, C.Q.; Xiong, Y.; Li, N.; Xu, J.; Liu, X.M.; Guo, Y.C. ‘Analysis of foodborne disease outbreaks in China mainland in 2013’. Chin. J. Food Hyg. 2018, 30, 293–298. [Google Scholar]
- Dong, P.; Zhu, L.; Mao, Y.; Liang, R.; Niu, L.; Zhang, Y.; Li, K.; Luo, X. Prevalence and profile of Salmonella from samples along the production line in Chinese beef processing plants. Food Control. 2014, 38, 54–60. [Google Scholar] [CrossRef]
- Cai, Y.; Tao, J.; Jiao, Y.; Fei, X.; Zhou, L.; Wang, Y.; Zheng, H.; Pan, Z.; Jiao, X. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 2016, 222, 56–64. [Google Scholar] [CrossRef]
- Jiang, Z.; Paudyal, N.; Xu, Y.; Deng, T.; Li, F.; Pan, H.; Peng, X.; He, Q.; Yue, M. Antibiotic resistance profiles of Salmonella, recovered from finishing pigs and slaughter facilities in Henan, China. Front. Microbiol. 2019, 10, 1513. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). ESBL-Producing Enterobacteriaceae in Healthcare Settings. Available online: https://www.cdc.gov/hai/organisms/ESBL.html (accessed on 18 April 2020).
- Vinueza-Burgos, C.; Ortega-Paredes, D.; Narvaez, C.; De Zutter, L.; Zurita, J. Characterization of cefotaxime resistant Escherichia coli isolated from broiler farms in ecuador. PLoS ONE 2019, 14, e0207567. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.R. Extended-Spectrum Beta-Lactamases Producing Escherichia coli (ESBLS-Producing, E. coli) in Dairy Farms in Beijing, China. Ph.D. Thesis, Chiang Mai University and Freie Universität Berlin, Berlin, Germany, 2013. [Google Scholar]
- Ali, T.; Ur Rahman, S.; Zhang, L.; Shahid, M.; Zhang, S.; Liu, G.; Gao, J.; Han, B. ESBL-producing Escherichia coli from cows suffering mastitis in China contain clinical class 1 integrons with CTX-M linked to ISCR1. Front. Microbiol. 2016, 7, 1931. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, S.; Shang, X.; Wang, X.; Wang, L.; Yan, Z.; Li, H. Prevalence and characteristics of extended spectrum β-lactamase-producing Escherichia coli from bovine mastitis cases in China. J. Integr. Agr. 2018, 17, 1246–1251. [Google Scholar] [CrossRef]
- Liu, G.; Ali, T.; Gao, J.; Ur, R.S.; Yu, D.; Barkema, H.W.; Huo, W.; Xu, S.; Shi, Y.; Kastelic, J.P.; et al. Co-occurrence of plasmid-mediated colistin resistance (mcr-1) and extended spectrum β-lactamase encoding genes in Escherichia coli from bovine mastitis milk in China. Microb. Drug Resist. 2019. [Google Scholar] [CrossRef]
- Pereira, R.; Williams, D.R.; Rossitto, P.; Adaska, J.; Okello, E.; Champagne, J.; Lehenbauer, T.W.; Li, X.; Chase, J.; Nguyen, T.; et al. Association between herd management practices and antimicrobial resistance in Salmonella spp. from cull dairy cattle in Central California. PeerJ 2019, 7, e6546. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S.; Bruini, I.; Magnani, R.; Cannistrà, N.; Brindani, F. Low prevalence of Salmonella enterica in cull dairy cattle at slaughter in Northern Italy. Ital. J. Food Saf. 2017, 6, 6172. [Google Scholar] [CrossRef] [PubMed]
- Abu Aboud, O.A.; Adaska, J.M.; Williams, D.R.; Rossitto, P.V.; Champagne, J.D.; Lehenbauer, T.W.; Atwill, R.; Li, X.; Aly, S.S. Epidemiology of Salmonella spp. In California cull dairy cattle: Prevalence of fecal shedding and diagnostic accuracy of pooled enriched broth culture of fecal samples. PeerJ 2016, 4, e2386. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Qu, D.; Zhang, X.; Shen, J.; Cui, S.; Shi, Y.; Xi, M.; Sheng., M.; Zhi, S.; Meng, J. Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int. J. Food Microbiol. 2010, 141, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, M.; Zhou, C.; Gu, G.; Liang, J.; Hou, X.; Wang, M.; Wei, P. Prevalence and antimicrobial resistance of retail-meat-borne Salmonella in southern China during the years 2009–2016: The diversity of contamination and the resistance evolution of multidrug-resistant isolates. Int. J. Food Microbiol. 2020, 333, 108790. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. Notice of the Ministry of Agriculture on Printing and Distributing the 13th Five-Year Development Plan for the National Herbivorous Animal Husbandry (2016–2020). Available online: http://www.moa.gov.cn/nybgb/2016/dibaqi/201712/t20171219_6102799.htm (accessed on 1 September 2020).
- Canadian Dairy Informational Center (CDIC). Breed Improvement and Genetic Evaluation. Culling and Replacement Rates in Dairy Herds in Canada. Available online: http://www.dairyinfo.gc.ca/index_e.php?s1=dff-fcil&s2=mrr-pcle&s3=cr-tr (accessed on 29 May 2020).
- Pinedo, P.J.; De Vries, A.; Webb, D.W. Dynamics of culling risk with disposal codes reported by dairy herd improvement dairy herds. J. Dairy Sci. 2010, 93, 2250–2261. [Google Scholar] [CrossRef]
- Stojkov, J.; Bowers, G.; Draper, M.; Duffield, T.; Duivenvoorden, P.; Groleau, M.; Haupstein, D.; Peters, R.; Pritchard, J.; Radom, C.; et al. Hot topic: Management of cull dairy cows—Consensus of an expert consultation in Canada. J. Dairy Sci. 2018, 101, 11170–11174. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.R.; Dohoo, I. Culling decisions of dairy farmers during a 3-year Salmonella control study. Prev. Vet. Med. 2011, 100, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhao, H.; Cui, S.; Wang, Y.; Xia, X.; Xi, M.; Wang, X.; Meng, J.; Ge, W. Prevalence and characterization of Salmonella enterica in dried milk-related infant foods in Shaanxi, China. J. Dairy Sci. 2014, 97, 6754–6760. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. Report on the Use of Veterinary Antibiotics of China in 2018. Off. Vet. Bull. 2019, 21, 57–59. [Google Scholar]
- Krömker, V.; Leimbach, S. Mastitis treatment—Reduction in antibiotic usage in dairy cows. Reprod. Domest. Anim. 2017, 52, 21–29. [Google Scholar] [CrossRef]
- Paudyal, N.; Pan, H.; Elbediwi, M.; Zhou, X.; Peng, X.; Li, X.; Fang, W.; Yue, M. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol. 2019, 19, 226–228. [Google Scholar] [CrossRef]
- Zhang, L.; Fu, Y.; Xiong, Z.; Ma, Y.; Wei, Y.; Qu, X.; Zhang, H.; Zhang, J.; Liao, M. Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front. Microbiol. 2018, 9, 2104. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1809. [Google Scholar] [CrossRef]
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. Announcement of the Ministry of Agriculture of the People’s Republic of China, No. 2428. Available online: http://www.moa.gov.cn/nybgb/2016/dibaqi/201712/t20171219_6102822.htm (accessed on 29 May 2020).
- Eguale, T.; Engidawork, E.; Gebreyes, W.A.; Asrat, D.; Alemayehu, H.; Medhin, G.; Johnson, R.P.; Gunn, J.S. Fecal prevalence, serotype distribution and antimicrobial resistance of Salmonellae in dairy cattle in central Ethiopia. BMC Microbiol. 2016, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Simova, V.; Voslarova, E.; Vecerek, V.; Passantino, A.; Bedanova, I. Effects of travel distance and season of the year on transport-related mortality in cattle. Anim. Sci. J. 2017, 88, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Dahl-Pedersen, K.; Herskin, M.S.; Houe, H.; Thomsen, P.T. Risk factors for deterioration of the clinical condition of cull dairy cows during transport to slaughter. Front. Vet. Sci. 2018, 5, 297. [Google Scholar] [CrossRef] [PubMed]
- Makino, S.; Kurazono, H.; Chongsanguam, M.; Hayashi, H.; Cheun, H.; Suzuki, S.; Shirahata, T. Establishment of the PCR system specific to Salmonella spp. and its application for the inspection of food and fecal samples. J. Vet. Med. Sci. 1999, 61, 1245–1247. [Google Scholar] [CrossRef]
- Zhai, L.; Kong, X.; Lu, Z.; Lv, F.; Zhang, C.; Bie, X. Detection of Salmonella enterica serovar Dublin by polymerase chain reaction in multiplex format. J. Microbiol. Methods 2014, 100, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, Y.; Shen, J.; Wu, C. Development of a novel hexa-plex PCR method for identification and serotyping of Salmonella species. Foodborne. Pathog. Dis. 2014, 11, 75. [Google Scholar] [CrossRef]
- Liu, B. Ming of molecular targets and development of multiplex PCR methods for serogouping and serotyping Salmonella spp. Ph.D. Thesis, Shanghai Jiao Tong University, Shanghai, China, 2012. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals: Informational Supplement, M31-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals: Twenty-Second Informational Supplement M100-S22; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Kwa, A.; Kasiakou, S.K.; Tam, V.H.; Falagas, M.E. Polymyxin B: Similarities to and differences from colistin (polymyxin E). Expert Rev. Anti Infect. Ther. 2007, 5, 811–821. [Google Scholar] [CrossRef]
- EURL-AR. Laboratory Protocol: Quantification of ESBL/AmpC-Producing Escherichia coli in Caecal Content and Fresh Meat Samples. European Union Reference Laboratory Antimicrobial Resistance. December 2017. Version 1. Available online: https://www.eurl-ar.eu/CustomerData/Files/Folders/21-protocols/399_esbl-ampc-quantification-protocol-19-03-2018.pdf (accessed on 20 June 2017).
- Hasman, H.; Mevius, D.; Veldman, K.; Olesen, I.; Aarestrup, F.M. Beta-lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in the Netherlands. J. Antimicrob. Chemother. 2005, 56, 115–121. [Google Scholar] [CrossRef]
- Rasheed, J.K.; Jay, C.; Metchock, B.; Berkowitz, F.; Weigel, L.; Crellin, J.; Steward, C.; Hill, B.; Medeiros, A.A.; Tenover, F.C. Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 1997, 41, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Dierikx, C.; van Essen-Zandbergen, A.; Veldman, K.; Smith, H.; Mevius, D. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet. Microbiol. 2010, 145, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Eurosurveillance 2016, 21, 30280. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).