Design and Characterization of a Novel Tool for the Antigenic Enrichment of Actinobacillus pleuropneumoniae Outer Membrane
Abstract
:1. Introduction
2. Results
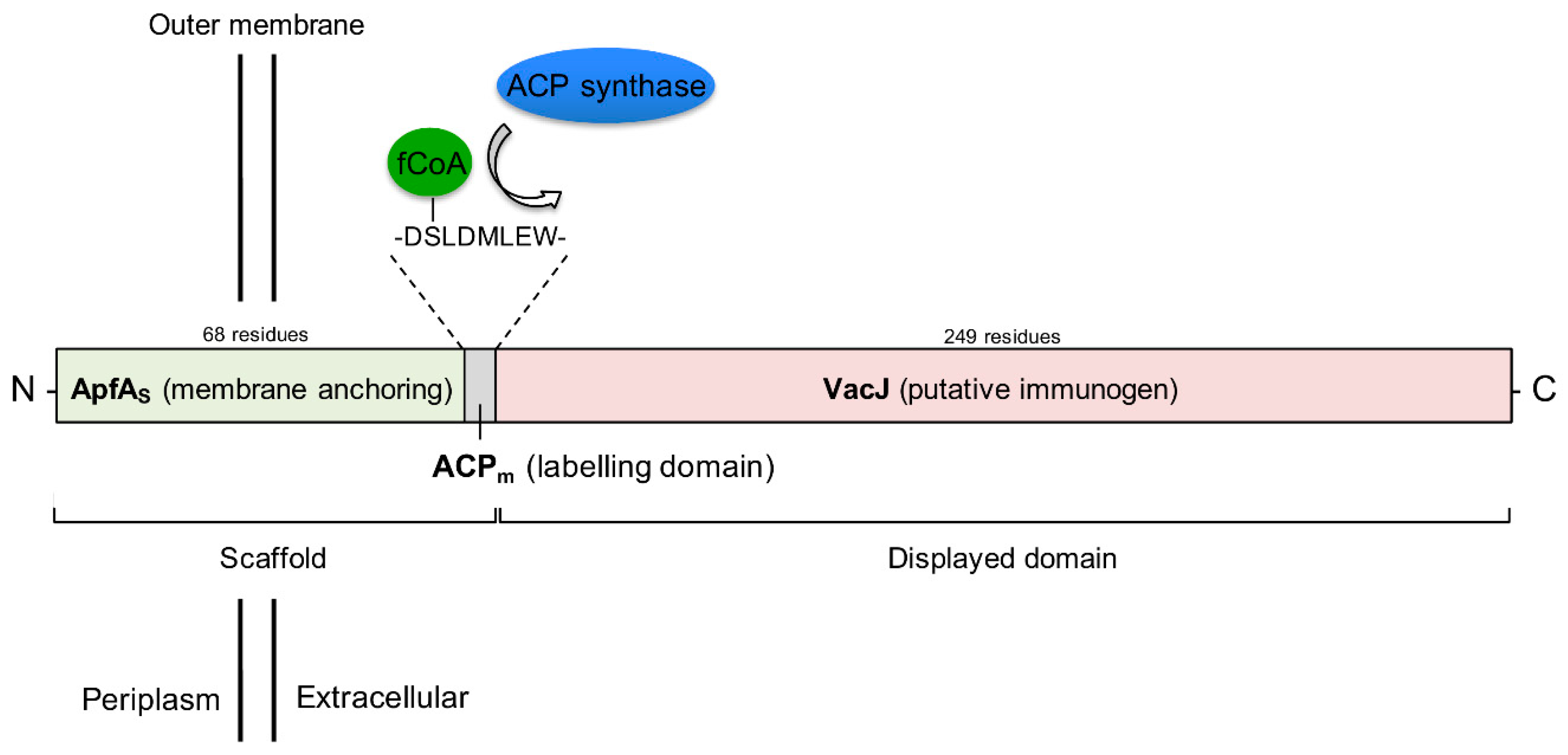
2.1. ApfAs is a Conserved OM Domain
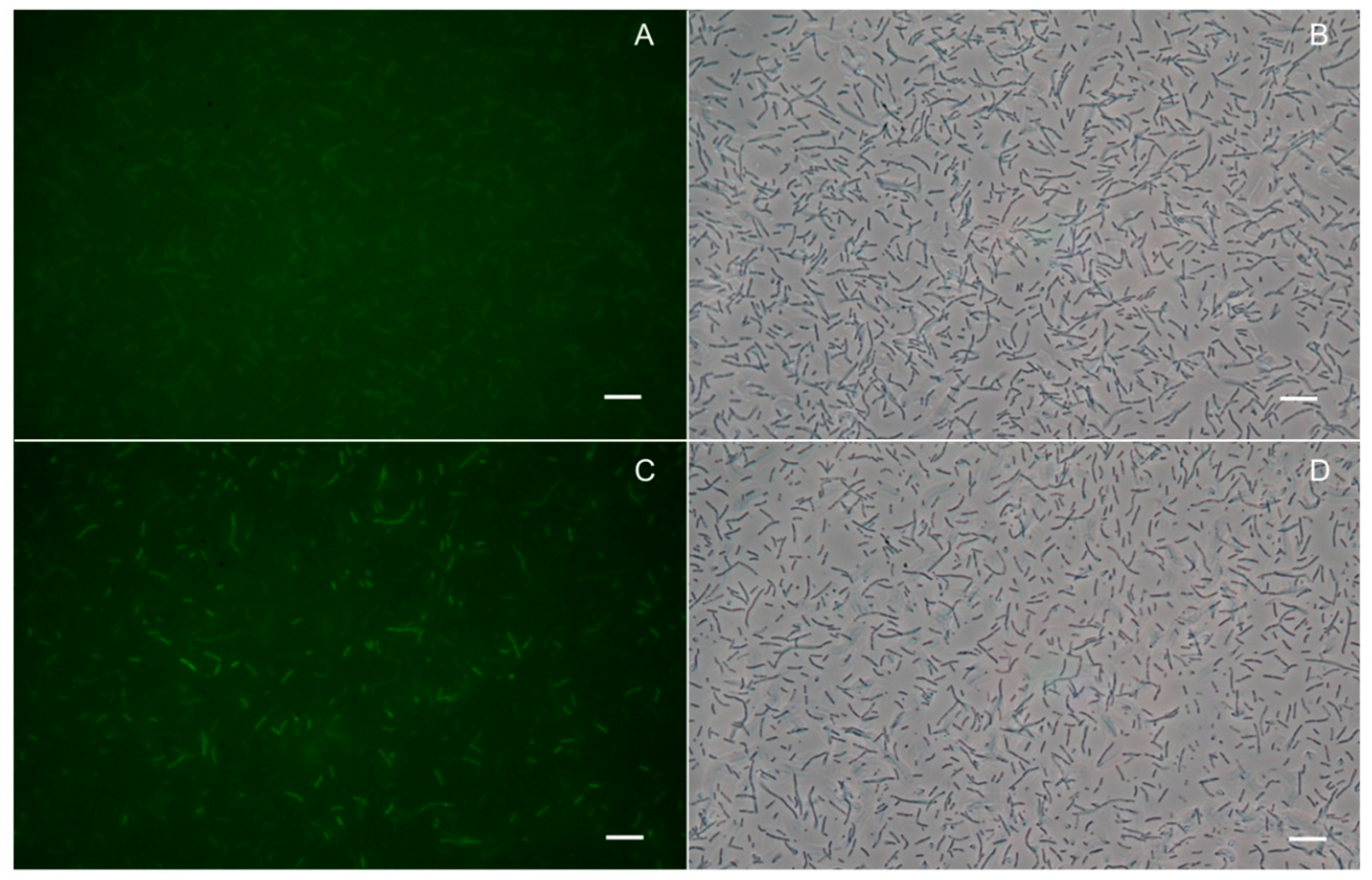
2.2. ApfAs-ACPm-VacJ is Expressed and Localized on A. pleuropneumoniae OM
3. Discussion
4. Materials and Methods
4.1. In Silico Functional Prediction of ApfA Domains
4.2. Design of the apfAs-ACPm-vacJ Chimera
4.3. Construction and Transfer of the apfAs-ACPm-vacJ Expression Vector
4.4. Outer Membrane Isolation
4.5. SDS-PAGE
4.6. Liquid Chromatography-Mass Spectrometry
4.7. Protein Identification by Scaffold Viewer
4.8. Flow Cytometry
4.9. Fluorescence Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Ullah, M.W.; Siddique, R.; Nabi, G.; Manan, S.; Yousaf, M.; Hou, H. Role of Recombinant DNA Technology to Improve Life. Int. J. Genom. 2016, 2016, 2405954. [Google Scholar] [CrossRef]
- Ellis, R.W.; Brodeur, B.R. New Bacterial Vaccines; Springer: Boston, MA, USA, 2003; ISBN 978-1-4613-4902-0. [Google Scholar]
- Flower, D.R.; Perrie, Y. Immunomic Discovery of Adjuvants and Candidate Subunit Vaccines; Springer: New York, NY, USA, 2013; ISBN 9781461450702. [Google Scholar]
- Tomar, N.; De, R.K. Immunoinformatics: A brief review. Methods Mol. Biol. 2014, 1184, 23–55. [Google Scholar]
- Sette, A. Reverse Vaccinology: Developing Vaccines in the Era of Genomics. Immunity 2010, 33, 530–541. [Google Scholar] [CrossRef] [Green Version]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Cenk Sahinalp, S.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Anderson, R.J.; Guru, S.; Weeratna, R.; Makinen, S.; Falconer, D.J.; Sheppard, N.C.; Lang, S.; Chang, B.; Goenaga, A.L.; Green, B.A.; et al. In vivo screen of genetically conserved Streptococcus pneumoniae proteins for protective immunogenicity. Vaccine 2016, 34, 6292–6300. [Google Scholar] [CrossRef]
- Lundberg, U.; Senn, B.M.; Schuler, W.; Meinke, A.; Hanner, M. Identification and characterization of antigens as vaccine candidates against Klebsiella pneumoniae. Hum. Vaccines Immunother. 2013, 9, 497–505. [Google Scholar] [CrossRef] [Green Version]
- Van Regenmortel, M.H.V. Structure-based reverse vaccinology failed in the case of HIV because it disregarded accepted immunological theory. Int. J. Mol. Sci. 2016, 17, 1591. [Google Scholar] [CrossRef] [Green Version]
- Donati, C.; Rappuoli, R. Reverse vaccinology in the 21st century: Improvements over the original design. Ann. N. Y. Acad. Sci. 2013, 1285, 115–132. [Google Scholar] [CrossRef]
- Gomes, A.; Byregowda, S.; Veeregowda, B.; Balamurugan, V. An Overview of Heterologous Expression Host Systems for the Production of Recombinant Proteins. Recomb. Gene Expr. 2016, 4, 15–51. [Google Scholar] [CrossRef] [Green Version]
- Bilgimol, J.C.; Suthakaran, P.; Sankaranarayanan, S.; Musti, M.; Kalimuthu, S.; Ganesan, M.; Sadananda, R.M. An overview of the parameters for recombinant protein expression in Escherichia coli. Cell Sci. Ther. 2015, 6, 1–7. [Google Scholar] [CrossRef]
- Jia, B.; Jeon, C.O. High-throughput recombinant protein expression in Escherichia coli: Current status and future perspectives. Open Biol. 2016, 6, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Thanassi, D.G.; Hultgren, S.J. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 2000, 12, 420–430. [Google Scholar] [CrossRef]
- Filloux, A. Secretion signal and protein targeting in Bacteria: A biological puzzle. J. Bacteriol. 2010, 192, 3847–3849. [Google Scholar] [CrossRef] [Green Version]
- Balbás, P.; Lorence, A. Recombinant Gene Expression Reviews and Protocols; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2004; Volume 267, ISBN 1592597742. [Google Scholar]
- Terpe, K. Overview of tag protein fusions: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003, 60, 523–533. [Google Scholar] [CrossRef]
- Lotze, J.; Reinhardt, U.; Seitz, O.; Beck-Sickinger, A.G. Peptide-tags for site-specific protein labelling in vitro and in vivo. Mol. BioSyst. 2016, 12, 1731–1745. [Google Scholar] [CrossRef] [Green Version]
- Yano, Y.; Matsuzaki, K. Tag–probe labeling methods for live-cell imaging of membrane proteins. Biochim. Biophys. Acta Biomembr. 2009, 1788, 2124–2131. [Google Scholar] [CrossRef] [Green Version]
- Botos, I.; Noinaj, N.; Buchanan, S.K. Insertion of proteins and lipopolysaccharide into the bacterial outer membrane. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160224. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Li, H.; Dong, H.; Zeng, Y.; Zhang, Z.; Paterson, N.G.; Stansfeld, P.J.; Wang, Z.; Zhang, Y.; Wang, W.; et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature 2016, 531, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Cho, B.K.; Knight, E.M.; Palsson, B. PCR-based tandem epitope tagging system for Escherichia coli genome engineering. Biotechniques 2006, 40, 67–72. [Google Scholar] [CrossRef]
- Bauler, L.D.; Hackstadt, T. Expression and Targeting of secreted proteins from Chlamydia trachomatis. J. Bacteriol. 2014, 196, 1325–1334. [Google Scholar] [CrossRef] [Green Version]
- Bossé, J.T.; Janson, H.; Sheehan, B.J.; Beddek, A.J.; Rycroft, A.N.; Simon Kroll, J.; Langford, P.R. Actinobacillus pleuropneumoniae: Pathobiology and pathogenesis of infection. Microbes Infect. 2002, 4, 225–235. [Google Scholar] [CrossRef]
- Ramjeet, M.; Deslandes, V.; Gouré, J.; Jacques, M. Actinobacillus pleuropneumoniae vaccines: From bacterins to new insights into vaccination strategies. Anim. Health Res. Rev. 2008, 9, 25–45. [Google Scholar] [CrossRef]
- Sjölund, M.; Wallgren, P. Field experience with two different vaccination strategies aiming to control infections with Actinobacillus pleuropneumoniae in a fattening pig herd. Acta Vet. Scand. 2010, 52, 23. [Google Scholar] [CrossRef] [Green Version]
- Sadilkova, L.; Nepereny, J.; Vrzal, V.; Sebo, P.; Osicka, R. Type IV fimbrial subunit protein ApfA contributes to protection against porcine pleuropneumonia. Vet. Res. 2012, 43, 2. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Li, L.; Chen, Z.; Yuan, H.; Chen, H.; Zhou, R. Adhesion Protein ApfA of Actinobacillus pleuropneumoniae Is Required for Pathogenesis and Is a Potential Target for Vaccine Development. Clin. Vaccine Immunol. 2013, 20, 287–294. [Google Scholar] [CrossRef] [Green Version]
- Shao, M.; Wang, Y.; Wang, C.; Guo, Y.; Peng, Y.; Liu, J.; Li, G.; Liu, H.; Liu, S. Evaluation of multicomponent recombinant vaccines against Actinobacillus pleuropneumoniae in mice. Acta Vet. Scand. 2010, 52, 52. [Google Scholar] [CrossRef] [Green Version]
- Deslandes, V.; Denicourt, M.; Girard, C.; Harel, J.; Nash, J.H.E.; Jacques, M. Transcriptional profiling of Actinobacillus pleuropneumoniae during the acute phase of a natural infection in pigs. BMC Genom. 2010, 11, 98. [Google Scholar] [CrossRef] [Green Version]
- Antenucci, F.; Fougeroux, C.; Bossé, J.T.; Magnowska, Z.; Roesch, C.; Langford, P.; Holst, P.J.; Bojesen, A.M. Identification and characterization of serovar-independent immunogens in Actinobacillus pleuropneumoniae. Vet. Res. 2017, 48, 74. [Google Scholar] [CrossRef] [Green Version]
- Antenucci, F.; Fougeroux, C.; Deeney, A.; Ørskov, C.; Rycroft, A.; Holst, P.J.; Bojesen, A.M. In vivo testing of novel vaccine prototypes against Actinobacillus pleuropneumoniae. Vet. Res. 2018, 49. [Google Scholar] [CrossRef] [Green Version]
- Tozakidis, I.E.P.; Sichwart, S.; Jose, J. Going beyond E. coli: Autotransporter based surface display on alternative host organisms. New Biotechnol. 2015, 32, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Hildegard, E.; Minh, D.B.; Schellack, C.; Nagy, E.; Meinke, A. Bacterial phage receptors, versatile tools for display of polypeptides on the cell surface. J. Bacteriol. 2001, 183, 6924–6935. [Google Scholar] [CrossRef] [Green Version]
- Chmielewski, M.; Kuehle, J.; Chrobok, D.; Riet, N.; Hallek, M.; Abken, H. FimH-based display of functional eukaryotic proteins on bacteria surfaces. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- George, N.; Pick, H.; Vogel, H.; Johnsson, N.; Johnsson, K. Specific labeling of cell surface proteins with chemically diverse compounds. J. Am. Chem. Soc. 2004, 126, 8896–8897. [Google Scholar] [CrossRef] [PubMed]
- McAllister, K.A.; Peery, R.B.; Zhao, G. Acyl carrier protein synthases from gram-negative, gram-positive, and atypical bacterial species: Biochemical and structural properties and physiological implications. J. Bacteriol. 2006, 188, 4737–4748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Koglin, A.; Wang, Y.; McMahon, A.P.; Walsh, C.T. An eight residue fragment of an acyl carrier protein suffices for post-translational introduction of fluorescent pantetheinyl arms in protein modification in vitro and in vivo. J. Am. Chem. Soc. 2008, 130, 9925–9930. [Google Scholar] [CrossRef] [Green Version]
- Roier, S.; Zingl, F.G.; Cakar, F.; Durakovic, S.; Kohl, P.; Eichmann, T.O.; Klug, L.; Gadermaier, B.; Weinzerl, K.; Prassl, R.; et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 2016, 7, 10515. [Google Scholar] [CrossRef] [Green Version]
- Kolberg, K.; Puettmann, C.; Pardo, A.; Fitting, J.; Barth, S. SNAP-Tag Technology: A General Introduction. Curr. Pharm. Des. 2013, 19, 5406–5413. [Google Scholar] [CrossRef]
- Smith, S.M. Strategies for the purification of membrane proteins. Methods Mol. Biol. 2011, 681, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Dong, Q.; Muruganujan, A.; Gaudet, P.; Lewis, S.; Thomas, P.D. PANTHER version 7: Improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2009, 38. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017, 44, D7–D19. [Google Scholar] [CrossRef] [Green Version]
- Gonzales, M.F.; Brooks, T.; Pukatzki, S.U.; Provenzano, D. Rapid protocol for preparation of electrocompetent Escherichia coli and Vibrio cholerae. J. Vis. Exp. 2013, 50684, e50684. [Google Scholar] [CrossRef] [Green Version]
- Bossé, J.T.; Durham, A.L.; Rycroft, A.N.; Kroll, J.S.; Langford, P.R. New Plasmid Tools for Genetic Analysis of Actinobacillus pleuropneumoniae and Other Pasteurellaceae. Appl. Environ. Microbiol. 2009, 75, 6124–6131. [Google Scholar] [CrossRef] [Green Version]
- Bossé, J.T.; Chaudhuri, R.R.; Li, Y.; Leanse, L.G.; Fernandez Crespo, R.; Coupland, P.; Holden, M.T.G.; Bazzolli, D.M.; Maskell, D.J.; Tucker, A.W.; et al. Complete Genome Sequence of MIDG2331, a Genetically Tractable Serovar 8 Clinical Isolate of Actinobacillus pleuropneumoniae. Genome Announc. 2016, 4, e01667-15. [Google Scholar] [CrossRef] [Green Version]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]


| Denomination | Sequence (5′-> 3′)/Accession Number (GenBank) | Function |
|---|---|---|
| App 5b L20 apfA gene | CP000569.1 | apfA ORF |
| App 5b L20 apfAs locus | Nucleotides 1-205 from A. pleuropneumoniae 5b L20 apfA | apfA stem |
| App 5b L20 vacJ gene | CP000569.1 | vacJ ORF |
| ACPm tag | GATTCGCTTGATATGCTGGAGTGG | Indirect detection of expression |
| pMK-express vector | GQ334690.1 | Naïve expression vector |
| pMK_apfas-ACPm-vacJ recombinant vector | Full sequence provided in supplementary Table S1 | Recombinant expression vector |
| apfA Fwd | CATcAGTAAAGGAGAATGCAgAAgCTAAGTCTTATTCGA | apfA amplification |
| apfA Rev | CCAGCATATCAAGCGAATCTCCGGTGTTATATATGCAGATCTCG | |
| vacJ Fwd | GCTTGATATGCTGGAGTGGAAgTTAAAgCAATTAAGgTTAGTAGCC | vacJ amplification |
| vacJ Rev | CAATTCACTGGCCGTTCTGCTCCTTTGCCCTATCC | |
| pMK Rev | ACTTAGcTTcTGCATTCTCCTTTACTgATGGTCAATTCTC | pMK vector linearization |
| pMK Fwd | GGGCAAAGGAGCAGAACGGCCAGTGAATTGTAATACG | |
| pMK Fwd sequencing | CGCCAACCGATAAAACCTAC | Upstream sequencing |
| vacJ Rev sequencing | CTTTTTACCCTCGCCCTCT | |
| vacJ Fwd sequencing | GGCGTGATTATGTGCCGA | Downstream sequencing |
| pMK Rev sequencing | CAATACGCAAACCGCCTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antenucci, F.; Ovsepian, A.; Wrobel, A.; Winther-Larsen, H.C.; Bojesen, A.M. Design and Characterization of a Novel Tool for the Antigenic Enrichment of Actinobacillus pleuropneumoniae Outer Membrane. Pathogens 2020, 9, 1014. https://doi.org/10.3390/pathogens9121014
Antenucci F, Ovsepian A, Wrobel A, Winther-Larsen HC, Bojesen AM. Design and Characterization of a Novel Tool for the Antigenic Enrichment of Actinobacillus pleuropneumoniae Outer Membrane. Pathogens. 2020; 9(12):1014. https://doi.org/10.3390/pathogens9121014
Chicago/Turabian StyleAntenucci, Fabio, Armen Ovsepian, Agnieszka Wrobel, Hanne Cecilie Winther-Larsen, and Anders Miki Bojesen. 2020. "Design and Characterization of a Novel Tool for the Antigenic Enrichment of Actinobacillus pleuropneumoniae Outer Membrane" Pathogens 9, no. 12: 1014. https://doi.org/10.3390/pathogens9121014
APA StyleAntenucci, F., Ovsepian, A., Wrobel, A., Winther-Larsen, H. C., & Bojesen, A. M. (2020). Design and Characterization of a Novel Tool for the Antigenic Enrichment of Actinobacillus pleuropneumoniae Outer Membrane. Pathogens, 9(12), 1014. https://doi.org/10.3390/pathogens9121014






