Correlation of NHR-48 Transcriptional Modulator Expression with Selected CYP Genes’ Expression during Thiabendazole Treatment of Anisakis simplex (s.l.)?—An In Vitro Study
Abstract
1. Introduction
2. Results
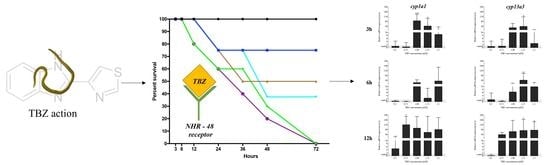
2.1. Survival of Larvae after TBZ Exposure
2.2. Bioinformatical Analyses
2.3. qReal-Time PCR
3. Discussion
4. Materials and Methods
4.1. Anisakis simplex (s.l.) Larvae
4.2. In Vitro Culture
4.3. Total RNA Isolation and cDNA Synthesis
4.4. Bioinformatical Analyses
4.5. qReal-Time PCR
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mattiucci, S.; Cipriani, P.; Levsen, A.; Paoletti, M.; Nascetti, G. Molecular Epidemiology of Anisakis and Anisakiasis: An Ecological and Evolutionary Road Map. Adv. Parasitol. 2018, 99, 93–263. [Google Scholar] [CrossRef]
- Valls, A.; Pascual, C.Y.; Martín Esteban, M. Anisakis allergy: An update. Rev. Fr. d’Allergol. d’Immunol. Clin. 2005, 45, 108–113. [Google Scholar] [CrossRef]
- Asaishi, K.; Nishino, C.; Ebata, T.; Totsuka, M.; Hayasaka, H.; Suzuki, T. Studies on the etiologic mechanism of anisakiasis. --1. Immunological reactions of digestive tract induced by Anisakis larva. Gastroenterol. Jpn. 1980, 15, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Sakanari, J.A.; McKerrow, J.H. Anisakiasis. Clin. Microbiol. Rev. 1989, 2, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Sakanari, J.A.; McKerrow, J.H. Identification of the Secreted Neutral Proteases from Anisakis simplex. J. Parasitol. 1990, 76, 625. [Google Scholar] [CrossRef]
- Mattiucci, S.; Fazii, P.; De Rosa, A.; Paoletti, M.; Megna, A.S.; Glielmo, A.; De Angelis, M.; Costa, A.; Meucci, C.; Calvaruso, V.; et al. Anisakiasis and Gastroallergic Reactions Associated with Anisakis pegreffii Infection, Italy. Emerg. Infect. Dis. 2013, 19, 496–499. [Google Scholar] [CrossRef]
- Audicana, M.T.; Kennedy, M.W. Anisakis simplex: From Obscure Infectious Worm to Inducer of Immune Hypersensitivity. Clin. Microbiol. Rev. 2008, 21, 360–379. [Google Scholar] [CrossRef]
- Audicana, M.T.; Ansotegui, I.J.; de Corres, L.F.; Kennedy, M.W. Anisakis simplex: Dangerous—Dead and alive? Trends Parasitol. 2002, 18, 20–25. [Google Scholar] [CrossRef]
- Moneo, I.; Caballero, M.L.; González-Muñoz, M.; Rodríguez-Mahillo, A.I.; Rodríguez-Perez, R.; Silva, A. Isolation of a heat-resistant allergen from the fish parasite Anisakis simplex. Parasitol. Res. 2005, 96, 285–289. [Google Scholar] [CrossRef]
- Bucci, C.; Gallotta, S.; Morra, I.; Fortunato, A.; Ciacci, C.; Iovino, P. Anisakis, just think about it in an emergency! Int. J. Infect. Dis. 2013, 17, e1071–e1072. [Google Scholar] [CrossRef]
- Dziekońska-Rynko, J.; Rokicki, J.; Jablonowski, Z. Effects of ivermectin and albendazole against Anisakis simplex in vitro and in guinea pigs. J. Parasitol. 2002, 88, 395–398. [Google Scholar] [CrossRef]
- Moore, D.A.; Girdwood, R.; Chiodini, P.L. Treatment of anisakiasis with albendazole. Lancet 2002, 360, 54. [Google Scholar] [CrossRef]
- Pacios, E.; Arias-Diaz, J.; Zuloaga, J.; Gonzalez-Armengol, J.; Villarroel, P.; Balibrea, J.L. Albendazole for the Treatment of Anisakiasis Ileus. Clin. Infect. Dis. 2005, 41, 1825–1826. [Google Scholar] [CrossRef] [PubMed]
- Ariasdiaz, J.; Zuloaga, J.; Vara, E.; Balibrea, J.; Balibrea, J. Efficacy of albendazole against Anisakis simplex larvae in vitro. Dig. Liver Dis. 2006, 38, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.C.; Navarro, M.C.; Martín-Sánchez, J.; Valero, A. Peppermint (Mentha piperita) and albendazole against anisakiasis in an animal model. Trop. Med. Int. Health 2014, 19, 1430–1436. [Google Scholar] [CrossRef]
- Hierro, I.; Valero, A.; Pérez, P.; González, P.; Cabo, M.M.; Montilla, M.P.; Navarro, M.C. Action of different monoterpenic compounds against Anisakis simplex s.l. L3 larvae. Phytomedicine 2004, 11, 77–82. [Google Scholar] [CrossRef]
- Lin, R.-J.; Chen, C.-Y.; Lee, J.-D.; Lu, C.-M.; Chung, L.-Y.; Yen, C.-M. Larvicidal Constituents of Zingiber officinale (Ginger) against Anisakis simplex. Planta Med. 2010, 76, 1852–1858. [Google Scholar] [CrossRef]
- Del Carmen Romero, M.; Valero, A.; Martín-Sánchez, J.; Navarro-Moll, M.C. Activity of Matricaria chamomilla essential oil against anisakiasis. Phytomedicine 2012, 19, 520–523. [Google Scholar] [CrossRef]
- Jones, L.M.; Flemming, A.J.; Urwin, P.E. NHR-176 regulates cyp-35d1 to control hydroxylation-dependent metabolism of thiabendazole in Caenorhabditis elegans. Biochem. J. 2015, 466, 37–44. [Google Scholar] [CrossRef]
- Borgers, M.; Nollin, S. De Ultrastructural Changes in Ascaris suum Intestine after Mebendazole Treatment In vivo. J. Parasitol. 1975, 61, 110. [Google Scholar] [CrossRef]
- Sakai, C.; Tomitsuka, E.; Esumi, H.; Harada, S.; Kita, K. Mitochondrial fumarate reductase as a target of chemotherapy: From parasites to cancer cells. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 643–651. [Google Scholar] [CrossRef]
- Shompole, S.; Yao, C.; Cheng, X.; Knox, D.; Johnson, S.; Jasmer, D.P. Distinct characteristics of two intestinal protein compartments discriminated by using fenbendazole and a benzimidazole resistant isolate of Haemonchus contortus. Exp. Parasitol. 2002, 101, 200–209. [Google Scholar] [CrossRef]
- Kotze, A.C.; Cowling, K.; Bagnall, N.H.; Hines, B.M.; Ruffell, A.P.; Hunt, P.W.; Coleman, G.T. Relative level of thiabendazole resistance associated with the E198A and F200Y SNPs in larvae of a multi-drug resistant isolate of Haemonchus contortus. Int. J. Parasitol. Drugs Drug Resist. 2012, 2, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Stuchlíková, L.R.; Matoušková, P.; Vokřál, I.; Lamka, J.; Szotáková, B.; Sečkařová, A.; Dimunová, D.; Nguyen, L.T.; Várady, M.; Skálová, L. Metabolism of albendazole, ricobendazole and flubendazole in Haemonchus contortus adults: Sex differences, resistance-related differences and the identification of new metabolites. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 50–58. [Google Scholar] [CrossRef]
- Laing, S.T.; Ivens, A.; Laing, R.; Ravikumar, S.; Butler, V.; Woods, D.J.; Gilleard, J.S. Characterization of the xenobiotic response of Caenorhabditis elegans to the anthelmintic drug albendazole and the identification of novel drug glucoside metabolites. Biochem. J. 2010, 432, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Prichard, R.K.; Hennessy, D.R.; Steel, J.W.; Lacey, E. Metabolite concentrations in plasma following treatment of cattle with five anthelmintics. Res. Vet. Sci. 1985, 39, 173–178. [Google Scholar] [CrossRef]
- Chukwudebe, A.C.; Wislocki, P.G.; Sanson, D.R.; Halls, T.D.J.; VandenHeuvel, W.J.A. Metabolism of Thiabendazole in Laying Hen and Lactating Goats. J. Agric. Food Chem. 1994, 42, 2964–2969. [Google Scholar] [CrossRef]
- Gonzalez, F.; Tukey, R. Drug metabolism. In Goodman & Gilman’s the Pharmacological Basis of Therapeutics; Parker, K., Ed.; McGraw-Hill Medical Publishing Division: New York, NY, USA, 2006; pp. 71–91. [Google Scholar]
- Zamanian, M.; Cook, D.E.; Zdraljevic, S.; Brady, S.C.; Lee, D.; Lee, J.; Andersen, E.C. Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Negl. Trop. Dis. 2018, 12, e0006368. [Google Scholar] [CrossRef]
- Stasiuk, S.J.; MacNevin, G.; Workentine, M.L.; Gray, D.; Redman, E.; Bartley, D.; Morrison, A.; Sharma, N.; Colwell, D.; Ro, D.K.; et al. Similarities and differences in the biotransformation and transcriptomic responses of Caenorhabditis elegans and Haemonchus contortus to five different benzimidazole drugs. Int. J. Parasitol. Drugs Drug Resist. 2019, 11, 13–29. [Google Scholar] [CrossRef]
- Jones, L.M.; Rayson, S.J.; Flemming, A.J.; Urwin, P.E. Adaptive and Specialised Transcriptional Responses to Xenobiotic Stress in Caenorhabditis elegans Are Regulated by Nuclear Hormone Receptors. PLoS ONE 2013, 8, e69956. [Google Scholar] [CrossRef]
- Chai, X.; Zeng, S.; Xie, W. Nuclear receptors PXR and CAR: Implications for drug metabolism regulation, pharmacogenomics and beyond. Expert Opin. Drug Metab. Toxicol. 2013, 9, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Ménez, C.; Alberich, M.; Courtot, E.; Guegnard, F.; Blanchard, A.; Aguilaniu, H.; Lespine, A. The transcription factor NHR-8: A new target to increase ivermectin efficacy in nematodes. PLoS Pathog. 2019, 15, e1007598. [Google Scholar] [CrossRef] [PubMed]
- Laing, R.; Bartley, D.J.; Morrison, A.A.; Rezansoff, A.; Martinelli, A.; Laing, S.T.; Gilleard, J.S. The cytochrome P450 family in the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2015, 45, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Mejía-Jaramillo, A.M.; Fernández, G.J.; Palacio, L.; Triana-Chávez, O. Gene expression study using real-time PCR identifies an NTR gene as a major marker of resistance to benznidazole in Trypanosoma cruzi. Parasit. Vectors 2011, 4, 169. [Google Scholar] [CrossRef]
- Kotze, A.C.; Hunt, P.W.; Skuce, P.; von Samson-Himmelstjerna, G.; Martin, R.J.; Sager, H.; Krücken, J.; Hodgkinson, J.; Lespine, A.; Jex, A.R.; et al. Recent advances in candidate-gene and whole-genome approaches to the discovery of anthelmintic resistance markers and the description of drug/receptor interactions. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 164–184. [Google Scholar] [CrossRef]
- Edwards, D.P. The role of coactivators and corepressors in the biology and mechanism of action of steroid hormone receptors. J. Mammary Gland Biol. Neoplasia 2000, 5, 307–324. [Google Scholar] [CrossRef]
- Magner, D.B.; Antebi, A. Caenorhabditis elegans nuclear receptors: Insights into life traits. Trends Endocrinol. Metab. 2008, 19, 153–160. [Google Scholar] [CrossRef]
- Ruaud, A.-F.; Bessereau, J.-L. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development 2006, 133, 2211–2222. [Google Scholar] [CrossRef]
- Holden-Dye, L.; Walker, R.J. How relevant is Caenorhabditis elegans as a model for the analysis of parasitic nematode biology? In Parasitic Helminths: Targets, Screens, Drugs and Vaccines; Caffrey, C.R., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2012; pp. 23–41. [Google Scholar]
- AlGusbi, S.; Krücken, J.; Ramünke, S.; von Samson-Himmelstjerna, G.; Demeler, J. Analysis of putative inhibitors of anthelmintic resistance mechanisms in cattle gastrointestinal nematodes. Int. J. Parasitol. 2014, 44, 647–658. [Google Scholar] [CrossRef]
- Menzel, R.; Rödel, M.; Kulas, J.; Steinberg, C.E.W. CYP35: Xenobiotically induced gene expression in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2005, 438, 93–102. [Google Scholar] [CrossRef]
- Chakrapani, B.P.S.; Kumar, S.; Subramaniam, J.R. Development and evaluation of an in vivo assay in Caenorhabditis elegans for screening of compounds for their effect on cytochrome P450 expression. J. Biosci. 2008, 33, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Albérich, M.; Ménez, C.; Sutra, J.-F.; Lespine, A. Ivermectin exposure leads to up-regulation of detoxification genes in vitro and in vivo in mice. Eur. J. Pharmacol. 2014, 740, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Lindblom, T.H.; Pierce, G.J.; Sluder, A.E. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Curr. Biol. 2001, 11, 864–868. [Google Scholar] [CrossRef]
- Iglesisas, L.; Valero, A.; Benitez, R.; Adroher, F.J. In vitro cultivation of Anisakis simplex: Pepsin increases survival and moulting from fourth larval to adult stage. Parasitology 2001, 123, 285–291. [Google Scholar] [CrossRef]
- Łopieńska-Biernat, E.; Paukszto, Ł.; Jastrzębski, J.P.; Myszczyński, K.; Polak, I.; Stryiński, R. Genome-wide analysis of Anisakis simplex sensu lato: The role of carbohydrate metabolism genes in the parasite’s development. Int. J. Parasitol. 2019, 49, 933–943. [Google Scholar] [CrossRef]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Łopieńska-Biernat, E.; Stryiński, R.; Paukszto, Ł.; Jastrzębski, J.P.; Makowczenko, K. The Selection of Reliable Reference Genes for RT-qPCR Analysis of Anisakis simplex Sensu Stricto Gene Expression from Different Developmental Stages. Acta Parasitol. 2020, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łopieńska-Biernat, E.; Stryiński, R.; Paukszto, Ł.; Jastrzębski, J.P. Correlation of NHR-48 Transcriptional Modulator Expression with Selected CYP Genes’ Expression during Thiabendazole Treatment of Anisakis simplex (s.l.)?—An In Vitro Study. Pathogens 2020, 9, 1030. https://doi.org/10.3390/pathogens9121030
Łopieńska-Biernat E, Stryiński R, Paukszto Ł, Jastrzębski JP. Correlation of NHR-48 Transcriptional Modulator Expression with Selected CYP Genes’ Expression during Thiabendazole Treatment of Anisakis simplex (s.l.)?—An In Vitro Study. Pathogens. 2020; 9(12):1030. https://doi.org/10.3390/pathogens9121030
Chicago/Turabian StyleŁopieńska-Biernat, Elżbieta, Robert Stryiński, Łukasz Paukszto, and Jan P. Jastrzębski. 2020. "Correlation of NHR-48 Transcriptional Modulator Expression with Selected CYP Genes’ Expression during Thiabendazole Treatment of Anisakis simplex (s.l.)?—An In Vitro Study" Pathogens 9, no. 12: 1030. https://doi.org/10.3390/pathogens9121030
APA StyleŁopieńska-Biernat, E., Stryiński, R., Paukszto, Ł., & Jastrzębski, J. P. (2020). Correlation of NHR-48 Transcriptional Modulator Expression with Selected CYP Genes’ Expression during Thiabendazole Treatment of Anisakis simplex (s.l.)?—An In Vitro Study. Pathogens, 9(12), 1030. https://doi.org/10.3390/pathogens9121030