Evaluation of West Nile Virus Diagnostic Capacities in Veterinary Laboratories of the Mediterranean and Black Sea Regions
Abstract
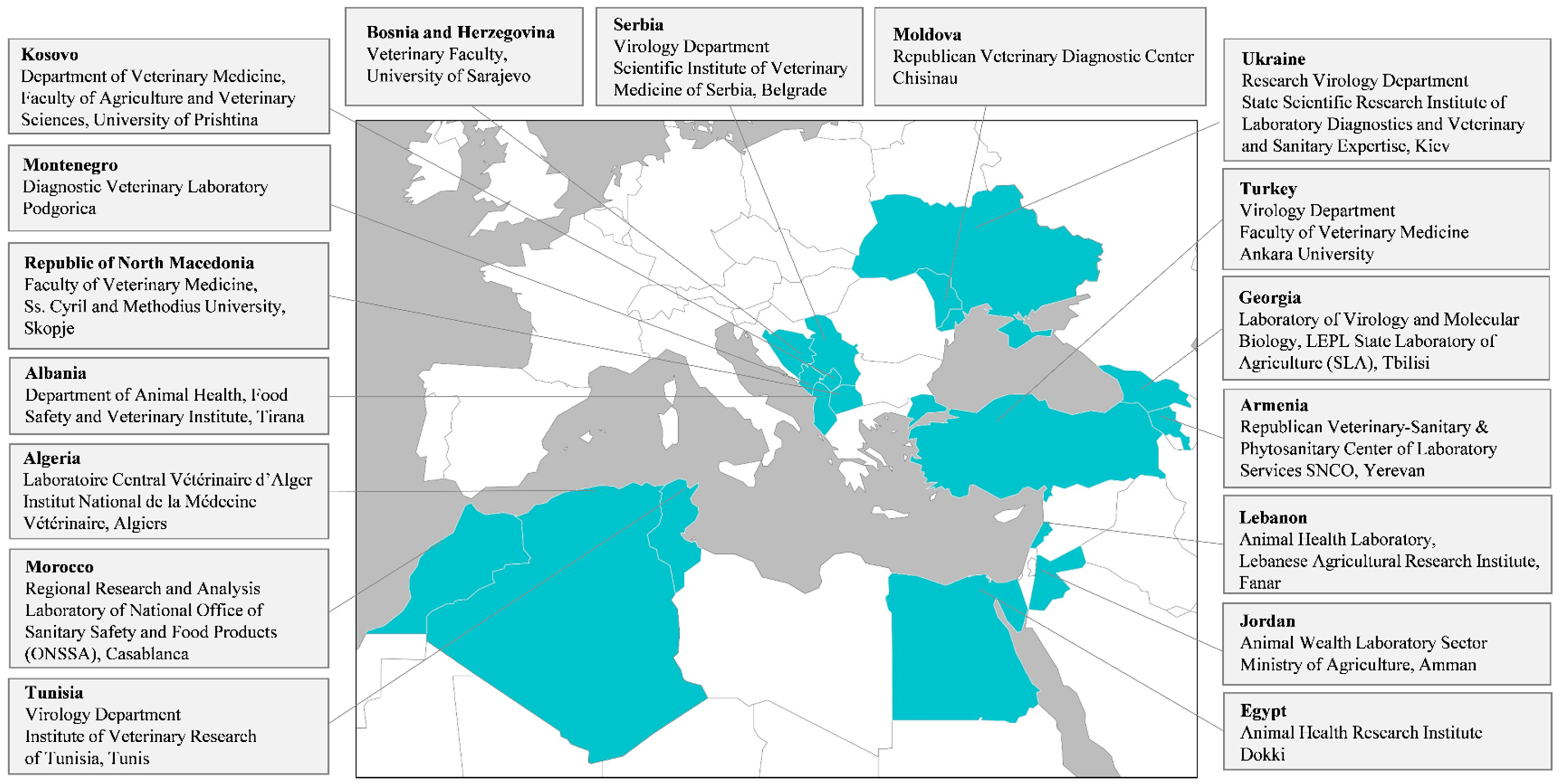
:1. Introduction
2. Results
2.1. Virus Genome Detection
2.2. Antibody Detection
3. Discussion
4. Materials and Methods
4.1. Call for Participation
4.2. Preparation of EQA Panel
4.2.1. Samples for Virus Genome Detection
4.2.2. Samples for Antibody Detection
4.3. EQA Details
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Statement
References
- Colpitts, T.M.; Conway, M.J.; Montgomery, R.R.; Fikrig, E. West Nile virus: Biology, transmission, and human infection. Clin. Microbiol. Rev. 2012, 25, 635–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLean, R.G.; Ubico, S.R.; Docherty, D.E.; Hansen, W.R.; Sileo, L.; McNamara, T.S. West Nile virus transmission and ecology in birds. Ann. N. Y. Acad. Sci. 2001, 951, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Chancey, C.; Grinev, A.; Volkova, E.; Rios, M. The global ecology and epidemiology of West Nile virus. BioMed Res. Int. 2015, 2015, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo-Olivares, J.; Mansfield, K.L.; Phipps, L.P.; Johnson, N.; Tearle, J.; Fooks, A.R. Antibody response in horses following experimental infection with West Nile virus lineages 1 and 2. Transbound. Emerg. Dis. 2011, 58, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ramírez, E.; Llorente, F.; Jiménez-Clavero, M.A. Experimental infections of wild birds with West Nile virus. Viruses 2014, 6, 752–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, C.; Jiménez-Clavero, M.A.; Leblond, A.; Durand, B.; Nowotny, N.; Leparc-Goffart, I.; Zientara, S.; Jourdain, E.; Lecollinet, S. Flaviviruses in Europe: Complex circulation patterns and their consequences for the diagnosis and control of West Nile disease. Int. J. Environ. Res. Public Health 2013, 10, 6049–6083. [Google Scholar] [CrossRef] [Green Version]
- Gossner, C.M.; Marrama, L.; Carson, M.; Allerberger, F.; Calistri, P.; Dilaveris, D.; Lecollinet, S.; Morgan, D.; Nowotny, N.; Paty, M.-C.; et al. West Nile virus surveillance in Europe: Moving towards an integrated animal-human-vector approach. Eurosurveillance 2017, 22, 30526. [Google Scholar] [CrossRef]
- ECDC. Epidemiological Update: West Nile virus Transmission Season in Europe. 2018. Available online: https://ecdc.europa.eu/en/news-events/epidemiological-update-west-nile-virus-transmission-season-europe-2018 (accessed on 12 October 2020).
- Clé, M.; Beck, C.; Salinas, S.; Lecollinet, S.; Gutiérrez, S.; Van de Perre, P.; Baldet, T.; Foulongne, V.; Simonin, Y. Usutu virus: A new threat? Epidemiol. Infect. 2019, 147, e232. [Google Scholar] [CrossRef] [Green Version]
- Weissenböck, H.; Kolodziejek, J.; Url, A.; Lussy, H.; Rebel-Bauder, B.; Nowotny, N. Emergence of Usutu virus, an African mosquito-borne flavivirus of the Japanese encephalitis virus group, Central Europe. Emerg. Infect. Dis. 2002, 8, 652–656. [Google Scholar] [CrossRef]
- Nikolay, B. A review of West Nile and Usutu virus co-circulation in Europe: How much do transmission cycles overlap? Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 609–618. [Google Scholar] [CrossRef] [Green Version]
- Durand, B.; Haskouri, H.; Lowenski, S.; Vachiery, N.; Beck, C.; Lecollinet, S. Seroprevalence of West Nile and Usutu viruses in military working horses and dogs, Morocco, 2012: Dog as an alternative WNV sentinel species? Epidemiol. Infect. 2016, 144, 1857–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Hassine, T.; De Massis, F.; Calistri, P.; Savini, G.; BelHaj Mohamed, B.; Ranen, A.; Di Gennaro, A.; Sghaier, S.; Hammami, S. First detection of co-circulation of West Nile and Usutu viruses in equids in the South-west of Tunisia. Transbound. Emerg. Dis. 2014, 61, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Kemenesi, G.; Buzás, D.; Zana, B.; Kurucz, K.; Krtinic, B.; Kepner, A.; Földes, F.; Jakab, F. First genetic characterization of Usutu virus from Culex pipiens mosquitoes Serbia, 2014. Infect. Genet. Evol. 2018, 63, 58–61. [Google Scholar] [CrossRef] [PubMed]
- del Amo, J.; Sotelo, E.; Fernández-Pinero, J.; Gallardo, C.; Llorente, F.; Agüero, M.; Jiménez-Clavero, M.A. A novel quantitative multiplex real-time RT-PCR for the simultaneous detection and differentiation of West Nile virus lineages 1 and 2, and of Usutu virus. J. Virol. Methods 2013, 189, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Llorente, F.; García-Irazábal, A.; Pérez-Ramírez, E.; Cano-Gómez, C.; Sarasa, M.; Vázquez, A.; Jiménez-Clavero, M.Á. Influence of flavivirus co-circulation in serological diagnostics and surveillance: A model of study using West Nile, Usutu and Bagaza viruses. Transbound. Emerg. Dis. 2019, 66, 2100–2106. [Google Scholar] [CrossRef] [PubMed]
- Riccardo, F.; Monaco, F.; Bella, A.; Savini, G.; Russo, F.; Cagarelli, R.; Dottori, M.; Rizzo, C.; Venturi, G.; Di Luca, M.; et al. An early start of West Nile virus seasonal transmission: The added value of One Heath surveillance in detecting early circulation and triggering timely response in Italy, June to July 2018. Eurosurveillance 2018, 23, 1800427. [Google Scholar] [CrossRef] [Green Version]
- Escadafal, C.; Gaayeb, L.; Riccardo, F.; Pérez-Ramírez, E.; Picard, M.; Dente, M.G.; Fernández-Pinero, J.; Manuguerra, J.-C.; Jiménez-Clavero, M.Á.; Declich, S.; et al. Risk of Zika virus transmission in the Euro-Mediterranean area and the added value of building preparedness to arboviral threats from a One Health perspective. BMC Public Health 2016, 16, 1219. [Google Scholar] [CrossRef] [Green Version]
- Dente, M.; Riccardo, F.; Nacca, G.; Ranghiasci, A.; Escadafal, C.; Gaayeb, L.; Jiménez-Clavero, M.Á.; Manuguerra, J.-C.; Picard, M.; Fernández-Pinero, J.; et al. Strengthening preparedness for arbovirus infections in Mediterranean and Black Sea countries: A conceptual framework to assess integrated surveillance in the context of the One Health strategy. Int. J. Environ. Res. Public Health 2018, 15, 489. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Ramírez, E.; Cano-Gómez, C.; Llorente, F.; Adzic, B.; Al Ameer, M.; Djadjovski, I.; El Hage, J.; El Mellouli, F.; Goletic, T.; Hovsepyan, H.; et al. External quality assessment of Rift Valley fever diagnosis in 17 veterinary laboratories of the Mediterranean and Black Sea regions. PLoS ONE 2020, 15, e0239478. [Google Scholar] [CrossRef]
- Eiden, M.; Vina-Rodríguez, A.; Hoffmann, B.; Ziegler, U.; Groschup, M.H. Two new real-time quantitative reverse transcription polymerase chain reaction assays with unique target sites for the specific and sensitive detection of lineages 1 and 2 West Nile virus strains. J. Vet. Diagn. Investig. 2010, 22, 748–753. [Google Scholar] [CrossRef] [Green Version]
- Weissenböck, H.; Bakonyi, T.; Chvala, S.; Nowotny, N. Experimental Usutu virus infection of suckling mice causes neuronal and glial cell apoptosis and demyelination. Acta Neuropathol. 2004, 108, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Niedrig, M.; Linke, S.; Zeller, H.; Drosten, C. First international proficiency study on West Nile virus molecular detection. Clin. Chem. 2006, 52, 1851–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linke, S.; MacKay, W.G.; Scott, C.; Wallace, P.; Niedrig, M. Second external quality assessment of the molecular diagnostic of West Nile virus: Are there improvements towards the detection of WNV? J. Clin. Virol. 2011, 52, 257–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niedrig, M.; Donoso Mantke, O.; Altmann, D.; Zeller, H. First international diagnostic accuracy study for the serological detection of West Nile virus infection. BMC Infect. Dis. 2007, 7, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchini, A.; Donoso-Mantke, O.; Papa, A.; Sambri, V.; Teichmann, A.; Niedrig, M. Second international diagnostic accuracy study for the serological detection of West Nile virus infection. PLoS Negl. Trop. Dis. 2013, 7, e2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reusken, C.; Baronti, C.; Mögling, R.; Papa, A.; Leitmeyer, K.; Charrel, R.N. Toscana, West Nile, Usutu and tick-borne encephalitis viruses: External quality assessment for molecular detection of emerging neurotropic viruses in Europe, 2017. Eurosurveillance 2019, 24, 1900051. [Google Scholar] [CrossRef] [Green Version]
- Pisani, G.; Pupella, S.; Cristiano, K.; Marino, F.; Simeoni, M.; Luciani, F.; Scuderi, G.; Sambri, V.; Rossini, G.; Gaibani, P.; et al. Detection of West Nile virus RNA (lineages 1 and 2) in an external quality assessment programme for laboratories screening blood and blood components for West Nile virus by nucleic acid amplification testing. Blood Transfus. 2012, 10, 515–520. [Google Scholar]
- Beck, C.; Lowenski, S.; Durand, B.; Bahuon, C.; Zientara, S.; Lecollinet, S. Improved reliability of serological tools for the diagnosis of West Nile fever in horses within Europe. PLoS Negl. Trop. Dis. 2017, 11, e0005936. [Google Scholar] [CrossRef]
- Sambri, V.; Capobianchi, M.R.; Cavrini, F.; Charrel, R.; Donoso-Mantke, O.; Escadafal, C.; Franco, L.; Gaibani, P.; Gould, E.A.; Niedrig, M.; et al. Diagnosis of West Nile virus human infections: Overview and proposal of diagnostic protocols considering the results of external quality assessment studies. Viruses 2013, 5, 2329–2348. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Triana, L.M.; Jeffries, C.L.; Mansfield, K.L.; Carnell, G.; Fooks, A.R.; Johnson, N. Emergence of West Nile virus lineage 2 in Europe: A review on the introduction and spread of a mosquito-borne disease. Front. Public Health 2014, 2, 271. [Google Scholar] [CrossRef] [Green Version]
- Napp, S.; Petrić, D.; Busquets, N. West Nile virus and other mosquito-borne viruses present in Eastern Europe. Pathog. Glob. Health 2018, 112, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Cadar, D.; Maier, P.; Müller, S.; Kress, J.; Chudy, M.; Bialonski, A.; Schlaphof, A.; Jansen, S.; Jöst, H.; Tannich, E.; et al. Blood donor screening for West Nile virus (WNV) revealed acute Usutu virus (USUV) infection, Germany, September 2016. Eurosurveillance 2017, 22, 30501. [Google Scholar] [CrossRef] [PubMed]
- Carletti, F.; Colavita, F.; Rovida, F.; Percivalle, E.; Baldanti, F.; Ricci, I.; De Liberato, C.; Rosone, F.; Messina, F.; Lalle, E.; et al. Expanding Usutu virus circulation in Italy: Detection in the Lazio region, central Italy, 2017 to 2018. Eurosurveillance 2019, 24, 1800649. [Google Scholar] [CrossRef] [PubMed]
- Scaramozzino, N.; Crance, J.-M.; Jouan, A.; DeBriel, D.; Stoll, F.; Garin, D. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J. Clin. Microbiol. 2001, 39, 1922–1927. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Clavero, M.A.; Sotelo, E.; Fernández-Pinero, J.; Llorente, F.; Blanco, J.M.; Rodriguez-Ramos, J.; Pérez-Ramírez, E.; Höfle, U. West Nile virus in Golden eagles, Spain, 2007. Emerg. Infect. Dis. 2008, 14, 1489–1491. [Google Scholar] [CrossRef] [Green Version]
- Barzon, L.; Pacenti, M.; Franchin, E.; Lavezzo, E.; Masi, G.; Squarzon, L.; Pagni, S.; Toppo, S.; Russo, F.; Cattai, M.; et al. Whole genome sequencing and phylogenetic analysis of West Nile virus lineage 1 and lineage 2 from human cases of infection, Italy, August 2013. Eurosurveillance 2013, 18, 20591. [Google Scholar] [CrossRef] [Green Version]
- Calzolari, M.; Chiapponi, C.; Bonilauri, P.; Lelli, D.; Baioni, L.; Barbieri, I.; Lavazza, A.; Pongolini, S.; Dottori, M.; Moreno, A. Co-circulation of two Usutu virus strains in Northern Italy between 2009 and 2014. Infect. Genet. Evol. 2017, 51, 255–262. [Google Scholar] [CrossRef]
- Pérez-Ramírez, E.; Llorente, F.; del Amo, J.; Nowotny, N.; Jiménez-Clavero, M.Á. Susceptibility and role as competent host of the red-legged partridge after infection with lineage 1 and 2 West Nile virus isolates of Mediterranean and Central European origin. Vet. Microbiol. 2018, 222, 39–45. [Google Scholar] [CrossRef]
- Sotelo, E.; Llorente, F.; Rebollo, B.; Camuñas, A.; Venteo, A.; Gallardo, C.; Lubisi, A.; Rodríguez, M.J.; Sanz, A.J.; Figuerola, J.; et al. Development and evaluation of a new epitope-blocking ELISA for universal detection of antibodies to West Nile virus. J. Virol. Methods 2011, 174, 35–41. [Google Scholar] [CrossRef]
| Virus | West Nile Virus (WNV) L1 | WNV L2 | Usutu Virus (USUV) | Japanese Encephalitis Virus (JEV) | WNV L1/ USUV | WNV L2/ USUV | -- | -- | |||
| Strain | SP07 | AUS08 | USU11 | Nakayama | SP07/ USU11 | AUS08/USU11 | -- | -- | |||
| Matrix (species) | Kidney (pheasant) | Liver (pheasant) | Serum (horse) | Blood (horse) | Serum (horse) | Heart (partridge) | Serum (horse) | Brain (horse) | Heart (pheasant) | ||
| Dilution | 10−2 | 10−3 | 10−1 | 10−3 | 10−3 | 10−2 | 10−4/10−4 | 10−2/10−4 | -- | -- | |
| Sample ID | W10 | W1 | W2 | W5 | W6 | W9 | W3 | W8 | W4 | W7 | |
| Reference value | Weak + | Weak + | Strong + | + | + | + | Weak + | Strong + | - | - | |
| Laboratory | % of correct results (by lab) | ||||||||||
| 1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | -- |
| 2 | - | - | + | + | + | + | - | + | - | - | 70 |
| 3 | + | - | + | + | + | + | - | + | - | - | 80 |
| 4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | -- |
| 5 | + | + | + | + | + | + | + | + | - | - | 100 |
| 6 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | -- |
| 7 | + | + | + | + | + | + | + | + | - | - | 100 |
| 8 | + | + | + | + | + | - | - | + | + | + | 60 |
| 9 | + | - | + | - | - | + | - | + | - | - | 60 |
| 10 | - | - | + | + | + | + | - | + | - | - | 70 |
| 11 | + | + | + | + | + | + | + | + | - | - | 100 |
| 12 | + | + | + | + | + | + | + | + | - | - | 100 |
| 13 | - | + | + | - | + | - | - | + | - | + | 50 |
| 14 | + | + | + | + | + | + | + | + | - | - | 100 |
| 15 | + | + | + | + | + | + | + | + | - | - | 100 |
| 16 | - | - | + | + | + | - | - | + | - | - | 60 |
| 17 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | -- |
| % of correct results (by sample) | 69.2 | 61.5 | 100 | 84.6 | 92.3 | 76.9 | 46.2 | 100 | 92.3 | 84.6 | |
| Virus | WNV L1 | WNV L2 | USUV | JEV | WNV L1/USUV | WNV L2/USUV | -- | -- | |||||
| Strain | SP07 | AUS08 | USU11 | Nakayama | SP07/ USU11 | AUS08/ USU11 | -- | -- | |||||
| Matrix (species) | Kidney (pheasant) | Liver (pheasant) | Serum (horse) | Blood (horse) | Serum (horse) | Heart (partridge) | Serum (horse) | Brain (horse) | Heart (pheasant) | ||||
| Dilution | 10−2 | 10−3 | 10−1 | 10−3 | 10−3 | 10−2 | 10−4/10−4 | 10−2/10−4 | -- | -- | |||
| Sample ID | W10 | W1 | W2 | W5 | W6 | W9 | W3 | W8 | W4 | W7 | |||
| Reference Ct value | WNV L1 (FAM) | 30.68 ± 0.88 | 33.60 ± 0.98 | No Ct | No Ct | No Ct | No Ct | 32.01 ± 0.41 | No Ct | No Ct | No Ct | ||
| WNV L2 (VIC) | No Ct | No Ct | 27.13 ± 0.57 | 34.28 ± 0.13 | No Ct | No Ct | No Ct | 31.34 ± 0.25 | No Ct | No Ct | |||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 29.24 ± 0.46 | No Ct | 32.1 ± 0.58 | 33.23 ± 0.31 | No Ct | No Ct | |||
| Lab | % of correct results for each virus (by lab) | % of correct overall results (by lab) † | |||||||||||
| 1 | WNV L1 (FAM) | No Ct | No Ct | 32.79 | No Ct | No Ct | No Ct | No Ct | No Ct | 31.98 | No Ct | 60 | 10 |
| WNV L2 (VIC) | No Ct | No Ct | No Ct | No Ct | 34.59 | 30 | 24.8 | No Ct | No Ct | No Ct | 40 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | No Ct | 38 | No Ct | No Ct | 33.53 | 36.2 | 50 | ||
| 2 | WNV L1 (FAM) | 27.75 | 33.32 | No Ct | No Ct | No Ct | No Ct | 32.4 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 37.04 | 32.87 | No Ct | No Ct | No Ct | 30.71 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 37.57 | No Ct | 34.1 | 36.19 | No Ct | No Ct | 100 | ||
| 3 | WNV L1 (FAM) | 30.15 | 31.71 | No Ct | No Ct | No Ct | No Ct | 30.31 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 21.66 | 26.66 | No Ct | No Ct | No Ct | 23.12 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 28.89 | No Ct | 28.58 | 30.24 | No Ct | No Ct | 100 | ||
| 4 | WNV L1 (FAM) | 28.28 | 31.07 | No Ct | No Ct | No Ct | No Ct | 30.44 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 24.53 | 32.55 | No Ct | No Ct | No Ct | 28.01 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 24.8 | No Ct | 27.45 | 28.02 | No Ct | No Ct | 100 | ||
| 5 | WNV L1 (FAM) | 29.44 | 32.74 | No Ct | No Ct | No Ct | No Ct | 30.52 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 24.67 | 32.65 | No Ct | No Ct | No Ct | 28.06 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 31.89 | No Ct | 36.92 | 33.74 | No Ct | No Ct | 100 | ||
| 6 | WNV L1 (FAM) | 31.6 | 36.8 | No Ct | No Ct | No Ct | No Ct | 34.5 | No Ct | No Ct | No Ct | 100 | 70 |
| WNV L2 (VIC) | No Ct | No Ct | 30.2 | 38.7 | No Ct | No Ct | No Ct | 33.9 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | 70 | ||
| 7 | WNV L1 (FAM) | 31.84 | 31.88 | No Ct | No Ct | No Ct | No Ct | 32.3 | No Ct | No Ct | No Ct | 100 | 70 |
| WNV L2 (VIC) | No Ct | No Ct | 25.54 | 32.78 | No Ct | No Ct | No Ct | 29.87 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | No Ct | 70 | ||
| 8 | WNV L1 (FAM) | 31.2 | No Ct | No Ct | No Ct | No Ct | No Ct | 33.46 | No Ct | No Ct | No Ct | 90 | 90 |
| WNV L2 (VIC) | No Ct | No Ct | 31 | 35.5 | No Ct | No Ct | No Ct | 32.5 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | - | ||
| 9 | WNV L1 (FAM) | 29 | 32 | No Ct | No Ct | No Ct | No Ct | 29 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 27 | 37 | No Ct | No Ct | No Ct | 32 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 28 | No Ct | 32 | 34 | No Ct | No Ct | 100 | ||
| 10 | WNV L1 (FAM) | 28.19 | 30.76 | No Ct | No Ct | No Ct | No Ct | 29.59 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 22.11 | 29.8 | No Ct | No Ct | No Ct | 26.78 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 28.39 | No Ct | 30.38 | 30.97 | No Ct | No Ct | 100 | ||
| 11 | WNV L1 (FAM) | 29.03 | 31.57 | No Ct | No Ct | No Ct | No Ct | 30.96 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 25.46 | 32.65 | No Ct | No Ct | No Ct | 28.75 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 28.81 | No Ct | 29.46 | 30.95 | No Ct | No Ct | 100 | ||
| 12 | WNV L1 (FAM) | 32.32 | 34.5 | No Ct | No Ct | No Ct | No Ct | 32.09 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 29.39 | 35.69 | No Ct | No Ct | No Ct | 32.11 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | - | ||
| 13 | WNV L1 (FAM) | 32.38 | 34.03 | No Ct | No Ct | No Ct | No Ct | 33.69 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 27.8 | 36.3 | No Ct | No Ct | No Ct | 31.45 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 36 | No Ct | 36.7 | 37.7 | No Ct | No Ct | 100 | ||
| 14 | WNV L1 (FAM) | 28 | 32 | No Ct | No Ct | No Ct | No Ct | 31.6 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 26.4 | 30.6 | No Ct | No Ct | No Ct | 26.4 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 30.8 | No Ct | 32.5 | 32.8 | No Ct | No Ct | 100 | ||
| 15 | WNV L1 (FAM) | 31.66 | 32.86 | No Ct | No Ct | No Ct | No Ct | 34.11 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 23.19 | 32.84 | No Ct | No Ct | No Ct | 27.41 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 29.45 | No Ct | 35.73 | 29.6 | No Ct | No Ct | 100 | ||
| 16 | WNV L1 (FAM) | 32 | 31.9 | No Ct | No Ct | No Ct | No Ct | 31.2 | No Ct | No Ct | No Ct | 100 | 100 |
| WNV L2 (VIC) | No Ct | No Ct | 24 | 33.5 | No Ct | No Ct | No Ct | 32.8 | No Ct | No Ct | 100 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 27.8 | No Ct | 32.6 | 28.2 | No Ct | No Ct | 100 | ||
| 17 | WNV L1 (FAM) | 34 | 30 | No Ct | No Ct | No Ct | No Ct | 32 | No Ct | No Ct | No Ct | 100 | 80 |
| WNV L2 (VIC) | No Ct | No Ct | 28 | 34 | No Ct | No Ct | No Ct | 37 | 32 | No Ct | 90 | ||
| USUV (Cy5) | No Ct | No Ct | No Ct | No Ct | 31 | No Ct | 28 | No Ct | No Ct | No Ct | 90 | ||
| % of correct results (by sample) ‡ | 94.1 | 88.2 | 94.1 | 94.1 | 82.3 | 94.1 | 82.3 | 76.4 | 94.1 | 94.1 | 94.1 | ||
| Serum Sample | Horse (past infection) | Horse (recent infection) | Non-infected horse | Negative commercial horse serum | |||||||
| W3 | W6 | W2 | W8 | W10 | W4 | W1 | W9 | W5 | W7 | ||
| Reference OD values | 0.11 ± 0.05 | 0.25 ± 0.21 | 0.34 ± 0.04 | 0.44 ± 0.09 | 0.48 ± 0.12 | 1.82 ± 0.21 | 0.07 ± 0.03 | 0.12 ± 0.11 | 0.10± 0.11 | 0.05 ± 0.03 | |
| Reference qualitative result | - | - | D/+ | + | + | + | - | - | - | - | |
| Laboratory | % of correct results (by lab) | ||||||||||
| 1 | - | - | + | - | + | + | - | - | - | - | 90 |
| 2 | - | - | D | + | + | + | - | - | - | - | 100 |
| 3 | - | - | + | + | + | + | - | - | - | - | 100 |
| 4 | - | - | + | + | + | - | - | - | - | - | 90 |
| 5 | - | - | + | + | + | + | - | - | - | - | 100 |
| 6 | - | - | - | - | D | + | - | - | - | - | 70 |
| 7 | - | - | + | + | + | + | - | - | - | - | 100 |
| 8 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | -- |
| 9 | - | - | + | + | + | + | - | - | - | - | 100 |
| 10 | - | - | D | - | + | + | - | - | - | - | 90 |
| 11 | - | - | + | + | + | + | - | - | - | - | 100 |
| 12 | - | - | + | + | + | + | - | - | - | - | 100 |
| 13 | - | - | + | + | + | + | - | - | - | - | 100 |
| 14 | - | - | D | D | + | + | - | - | - | - | 90 |
| 15 | - | D | D | + | + | + | - | - | - | - | 90 |
| 16 | - | - | D | + | + | + | - | - | - | - | 100 |
| 17 | - | - | - | D | + | + | - | - | - | - | 80 |
| % of correct results (by sample) | 100 | 93.7 | 87.5 | 68.7 | 93.7 | 93.7 | 100 | 90 | 100 | 100 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Ramírez, E.; Cano-Gómez, C.; Llorente, F.; Vodica, A.; Veljović, L.; Toklikishvilli, N.; Sherifi, K.; Sghaier, S.; Omani, A.; Kustura, A.; et al. Evaluation of West Nile Virus Diagnostic Capacities in Veterinary Laboratories of the Mediterranean and Black Sea Regions. Pathogens 2020, 9, 1038. https://doi.org/10.3390/pathogens9121038
Pérez-Ramírez E, Cano-Gómez C, Llorente F, Vodica A, Veljović L, Toklikishvilli N, Sherifi K, Sghaier S, Omani A, Kustura A, et al. Evaluation of West Nile Virus Diagnostic Capacities in Veterinary Laboratories of the Mediterranean and Black Sea Regions. Pathogens. 2020; 9(12):1038. https://doi.org/10.3390/pathogens9121038
Chicago/Turabian StylePérez-Ramírez, Elisa, Cristina Cano-Gómez, Francisco Llorente, Ani Vodica, Ljubiša Veljović, Natela Toklikishvilli, Kurtesh Sherifi, Soufien Sghaier, Amel Omani, Aida Kustura, and et al. 2020. "Evaluation of West Nile Virus Diagnostic Capacities in Veterinary Laboratories of the Mediterranean and Black Sea Regions" Pathogens 9, no. 12: 1038. https://doi.org/10.3390/pathogens9121038
APA StylePérez-Ramírez, E., Cano-Gómez, C., Llorente, F., Vodica, A., Veljović, L., Toklikishvilli, N., Sherifi, K., Sghaier, S., Omani, A., Kustura, A., Krstevski, K., Karayel-Hacioglu, I., Hagag, N. M., El Hage, J., Davdyan, H., Bintarif, M. S., Adzic, B., Abouchoaib, N., Jiménez-Clavero, M. Á., & Fernández-Pinero, J. (2020). Evaluation of West Nile Virus Diagnostic Capacities in Veterinary Laboratories of the Mediterranean and Black Sea Regions. Pathogens, 9(12), 1038. https://doi.org/10.3390/pathogens9121038










