Potential of Bioremediation and PGP Traits in Streptomyces as Strategies for Bio-Reclamation of Salt-Affected Soils for Agriculture
Abstract
:1. Introduction
1.1. The Genus Streptomyces
1.2. Streptomyces Life Cycle
1.3. Streptomyces Applications
2. Streptomyces in Bioremediation
2.1. Microbial Mechanisms Used for Bioremediation
2.2. The Case of Boron-Mining Environmental Impact
3. Streptomyces in Plant Growth Promotion
3.1. PGP Streptomyces against Biotic Stressors
3.2. PGP Streptomyces against Abiotic Stressors
4. Bio-Reclamation of Saline Soils
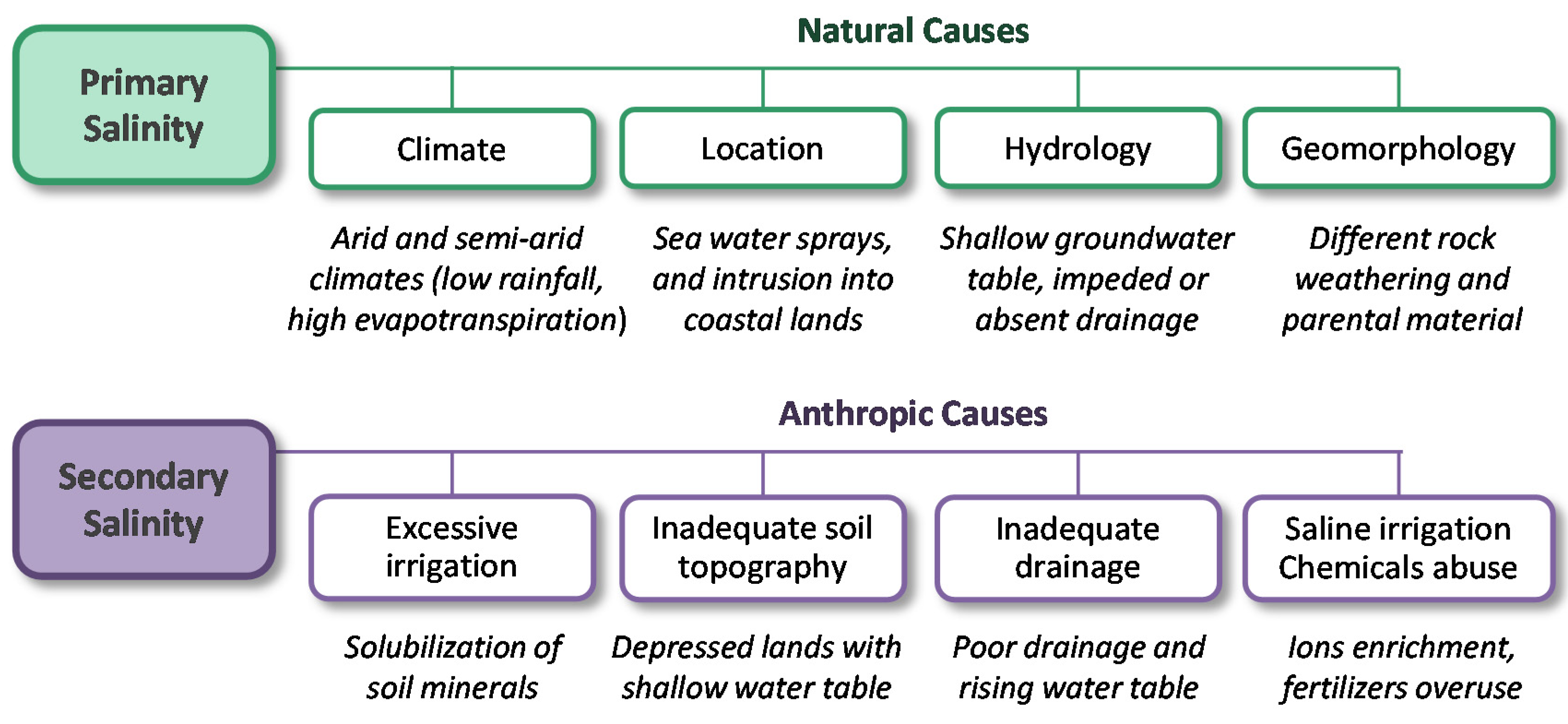
4.1. Soil Salinity, Causes and Effects
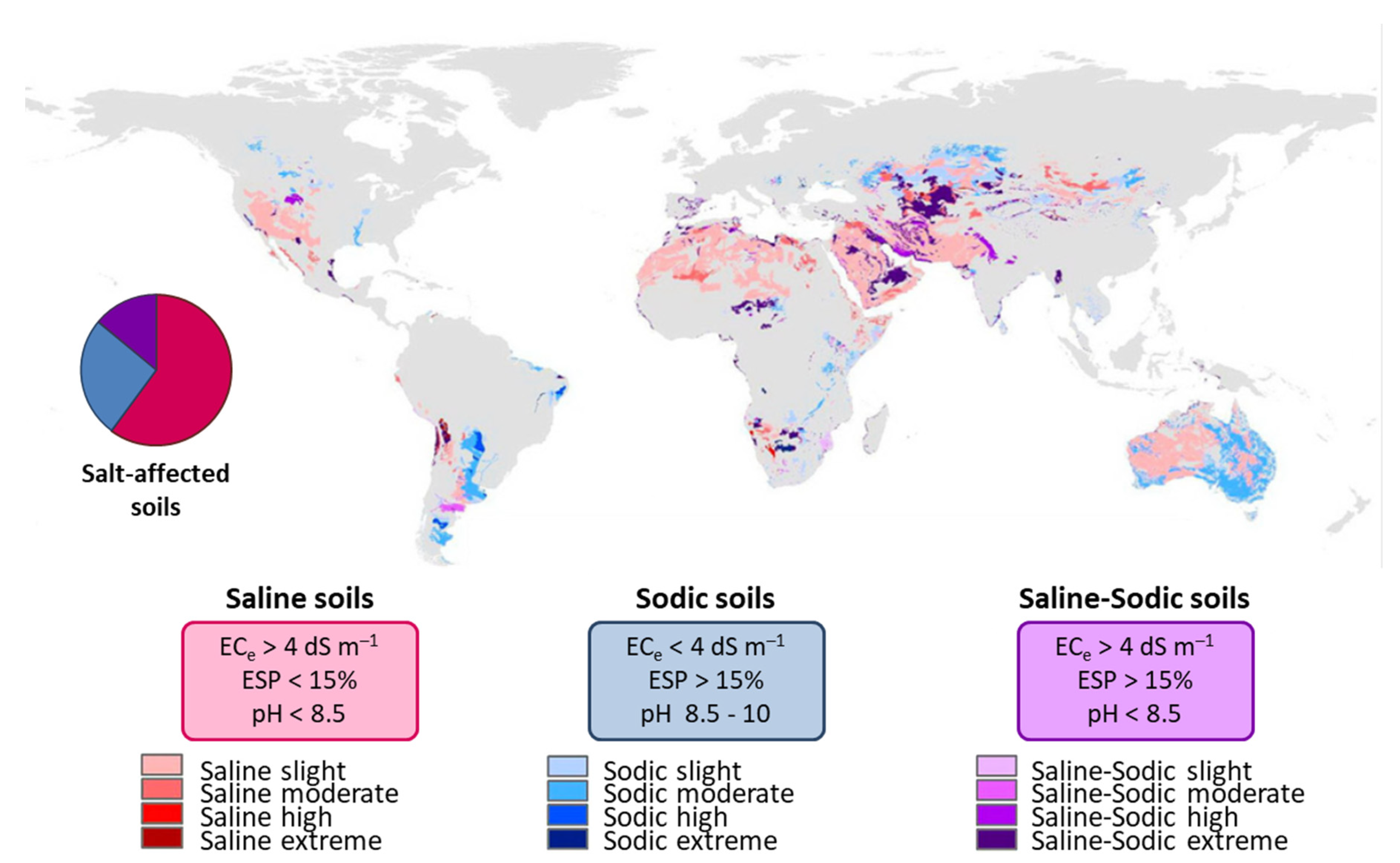
4.2. Salt-Affected Soils Classification and Distribution
4.3. Reclamation vs. Bio-Reclamation of Salt-Affected Soils
4.4. Streptomyces in Salt-Affected Soils
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Alexander, D. Bacteria and Archea. In Principles and Applications of Soil Microbiology; Sylvia, D., Fuhrmann, J., Hartel, P., Zuberer, D., Eds.; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2005; pp. 101–139. [Google Scholar]
- Atlas, R.M.; Bartha, R. Ecología Microbiana y Microbiología Ambiental, 4th ed.; Addison Wesley: Madrid, Spain, 1998. [Google Scholar]
- Zuberer, D.A.; Wollum, A.G. Introduction and historical perspective. In Principles and Applications of Soil Microbiology; Sylvia, D., Fuhrmann, J., HArtel, P., Zuberer, D., Eds.; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2005; pp. 3–25. [Google Scholar]
- Matsukawa, E.; Nakagawa, Y.; Iimura, Y.; Hayakawa, M. A new enrichment method for the selective isolation of streptomycetes from the root surfaces of herbaceous plants. Actinomycetologica 2007, 21, 66–69. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Li, H.; Pan, Y.; Wang, L.; Tan, H.; Liu, G. SCO3129, a TetR family regulator, is responsible for osmotic stress in Streptomyces coelicolor. Synth. Syst. Biotechnol. 2018, 3, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Moraga, N.B.; Irazusta, V.; Amoroso, M.J.; Rajal, V.B. Bio-precipitates produced by two autochthonous boron tolerant Streptomyces strains. J. Environ. Chem. Eng. 2017, 5, 3373–3383. [Google Scholar] [CrossRef] [PubMed]
- Lacey, J. Actinomycetes in soils, composts and fodders. In Actinomycetales: Characteristics and Practical Importance; Sykes, G., Skinner, F.A., Eds.; Academic Press: London, UK, 1973; pp. 231–251. [Google Scholar]
- Elander, R.P. Microbial screening, selection and strain improvement. In Basic Biotechnology; Bu Lock, J., Kristiansen, B., Eds.; Academic Press: London, UK, 1987; pp. 217–251. [Google Scholar]
- Keiser, T.; Viv, M.J.; Buttner, M.J.; Chater, K.F. Preparation and analysis of genomic plasmid DNA. In Practical Streptomyces Genetics; Keiser, T., Viv, M.J., Buttner, M.J., Chater, K.F., Hopwood, D.A., Eds.; The John Innes Foundation: Norwich, UK, 2000; pp. 161–210. [Google Scholar]
- Goodfellow, M.; Mordaski, M.; Williams, S. The biology of actinomycetes. In Extremophiles: Microbial Life in Extreme Environments; Horikoshi, K., Grant, W., Eds.; Academic Press: Orlando, FL, USA, 1984. [Google Scholar]
- Tokala, R.K.; Strap, J.L.; Jung, C.M.; Crawford, D.L.; Salove, M.H.; Deobald, L.A.; Bailey, J.F.; Morra, M.J. Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the Pea Plant (Pisum sativum). Appl. Environ. Microbiol. 2002, 68, 2161–2171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldesuquy, H.S.; Mansour, F.A.; Abo-Hamed, S.A. Effect of the culture filtrates of Streptomyces on growth and productivity of wheat plants. Folia Microbiol. (Praha) 1998, 43, 465–470. [Google Scholar] [CrossRef]
- Sadeghi, A.; Karimi, E.; Dahaji, P.A.; Javid, M.G.; Dalvand, Y.; Askari, H. Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World J. Microbiol. Biotechnol. 2012, 28, 1503–1509. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Srinivas, V.; Alekhya, G.; Prakash, B.; Kudapa, H.; Rathore, A.; Varshney, R.K. The extent of grain yield and plant growth enhancement by plant growth-promoting broad-spectrum Streptomyces sp. in chickpea. Springerplus 2015, 4, 31. [Google Scholar] [CrossRef] [Green Version]
- Gopalakrishnan, S.; Srinivas, V.; Vidya, M.S.; Rathore, A. Plant growth-promoting activities of Streptomyces spp. in sorghum and rice. Springer Plus 2013, 2, 574. [Google Scholar] [CrossRef] [Green Version]
- Gopalakrishnan, S.; Vadlamudi, S.; Bandikinda, P.; Sathya, A.; Vijayabharathi, R.; Rupela, O.; Kudapa, H.; Katta, K.; Varshney, R.K. Evaluation of Streptomyces strains isolated from herbal vermicompost for their plant growth-promotion traits in rice. Microbiol. Res. 2014, 169, 40–48. [Google Scholar] [CrossRef] [Green Version]
- Hernández, A.; Rives, N.; Caballero, A.; Hernández, A.; Heydrich, M. Caracterización de rizobacterias asociadas al cultivo del maíz en la producción de metabolitos del tipo AIA, sideróforos y ácido salicílico. Rev. Colomb. Biotecnol. 2004, VI, 6–13. [Google Scholar]
- Ravel, J.; Amoroso, M.J.; Colwell, R.R.; Hill, R.T. Mercury resistant actinomycetes form Chesapeake Bay. FEMS Microbiol. Lett. 1998, 162, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Watve, M.G.; Tickoo, R.; Jog, M.M.; Bhole, B.D. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Alanis, A.J. Resistance to antibiotics: Are we in the post-antibiotic era? Arch. Med. Res. 2005, 36, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Omura, S. Isolation of new actinomycete strains for the screening of new bioactive compounds. J. Gen. Appl. Microbiol. 2003, 49, 141–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cañaveras, J.C.; Hoyos, M.; Sanchez-Moral, S.; Sanz-Rubio, E.; Bedoya, J.; Soler, V.; Groth, I.; Schumann, P.; Laiz, L.; Gonzalez, I.; et al. Microbial communities associated with hydromagnesite and needle-fiber aragonite deposits in a karstic cave (Altamira, Northern Spain). Geomicrobiol. J. 1999, 16, 9–25. [Google Scholar]
- Haferburg, G.; Kloess, G.; Schmitz, W.; Kothe, E. “Ni-struvite”—A new biomineral formed by a nickel resistant Streptomyces acidiscabies. Chemosphere 2008, 72, 517–523. [Google Scholar] [CrossRef]
- Flärdh, K. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 2003, 6, 564–571. [Google Scholar] [CrossRef]
- Chater, K.F. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 1972, 72, 9–28. [Google Scholar] [CrossRef] [Green Version]
- Claessen, D.; Rozen, D.E.; Kuipers, O.P.; Søgaard-Andersen, L.; van Wezel, G.P. Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 2014, 12, 115. [Google Scholar] [CrossRef] [Green Version]
- Elliot, M.A.; Buttner, M.J.; Nodwell, J.R. Multicellular development in Streptomyces. In Myxobacteria; Whitworth, D.E., Ed.; American Society of Microbiology: Washington, DC, USA, 2008; pp. 419–438. ISBN 9781555814205. [Google Scholar]
- Wildermuth, H.; Hopwood, D.A. Septation during sporulation in Streptomyces coelicolor. J. Gen. Microbiol. 1970, 60, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Jakimowicz, D.; van Wezel, G.P. Cell division and DNA segregation in Streptomyces: How to build a septum in the middle of nowhere? Mol. Microbiol. 2012, 85, 393–404. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, B.; Chen, M.; Crawford, R.; Ivanova, E. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 2009, 14, 2535–2554. [Google Scholar] [CrossRef] [PubMed]
- Sheng, G.; Yu, H.; Yue, Z. Production of extracellular polymeric substances from Rhodopseudomonas acidophila in presence of toxic substances. Appl. Microbiol. Biotechnol. 2005, 69, 216–222. [Google Scholar] [CrossRef]
- Mendez, C.; Braña, A.F.; Manzanal, M.B.; Hardisson, C. Role of substrate mycelium in colony development in Streptomyces. Can. J. Microbiol. 1985, 31, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, H. Development and organization of the aerial mycelium in Streptomyces coelicolor. J. Gen. Microbiol. 1970, 60, 43–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibb, M.J. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 2005, 8, 208–215. [Google Scholar] [CrossRef]
- van Wezel, G.P.; McDowall, K.J. The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Nat. Prod. Rep. 2011, 28, 1311–1333. [Google Scholar] [CrossRef]
- Manteca, A.; Ye, J.; Sánchez, J.; Jensen, O.N. Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J. Proteome Res. 2011, 10, 5481–5492. [Google Scholar] [CrossRef]
- Bibb, M.J.; Domonkos, A.; Chandra, G.; Buttner, M.J. Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by sigma(BldN) and a cognate anti-sigma factor, RsbN. Mol. Microbiol. 2012, 84, 1033–1049. [Google Scholar] [CrossRef]
- Keijser, B.J.F.; Noens, E.E.E.; Kraal, B.; Koerten, H.K.; van Wezel, G.P. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol. Lett. 2003, 225, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Claessen, D.; Rink, R.; de Jong, W.; Siebring, J.; de Vreugd, P.; Boersma, F.G.H.; Dijkhuizen, L.; Wosten, H.A.B. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003, 17, 1714–1726. [Google Scholar] [CrossRef] [Green Version]
- Wildermuth, H.; Wehrli, E.; Horne, R.W. The surface structure of spores and aerial mycelium in Streptomyces coelicolor. J. Ultrastruct. Res. 1971, 35, 168–180. [Google Scholar] [CrossRef]
- Claessen, D.; de Jong, W.; Dijkhuizen, L.; Wösten, H.A.B. Regulation of Streptomyces development: Reach for the sky! Trends Microbiol. 2006, 14, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Wösten, H.A.; van Wetter, M.A.; Lugones, L.G.; van der Mei, H.C.; Busscher, H.J.; Wessels, J.G. How a fungus escapes the water to grow into the air. Curr. Biol. 1999, 9, 85–88. [Google Scholar] [CrossRef] [Green Version]
- Dragos, A.; Kovacs, A.T.; Claessen, D. The role of functional amyloids in multicellular growth and development of Gram-positive bacteria. Biomolecules 2017, 7, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flärdh, K.; Leibovitz, E.; Buttner, M.J.; Chater, K.F. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol. Microbiol. 2000, 38, 737–749. [Google Scholar] [CrossRef]
- Celler, K.; Koning, R.I.; Koster, A.J.; van Wezel, G.P. Multidimensional view of the bacterial cytoskeleton. J. Bacteriol. 2013, 195, 1627–1636. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.E.; Ho, L.; Rees, C.A.; Hill, J.E.; Nodwell, J.R.; Elliot, M.A. Streptomyces exploration is triggered by fungal interactions and volatile signals. Elife 2017, 6, e21738. [Google Scholar] [CrossRef]
- Schatz, A.; Bugie, E.; Waksman, S.A. Streptomycin, a substance exhibiting antibiotic activity against Gram-Positive and Gram-Negative bacteria. Proc. Soc. Exp. Biol. Med. 1944, 55, 66–69. [Google Scholar] [CrossRef]
- Elsevier, B.V. ScienceDirect. Available online: https://www.sciencedirect.com/ (accessed on 10 October 2019).
- Jones, S.E.; Elliot, M.A. Streptomyces exploration: Competition, volatile communication and new bacterial behaviours. Trends Microbiol. 2017, 25, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A. Highlights of Streptomyces genetics. Heredity (Edinb.) 2019, 123, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.F.; Teixeira, M.F.S.; Converti, A.; Porto, A.L.F.; Sarubbo, L.A. Production of a new lipoprotein biosurfactant by Streptomyces sp. DPUA1566 isolated from lichens collected in the Brazilian Amazon using agroindustry wastes. Biocatal. Agric. Biotechnol. 2019, 17, 142–150. [Google Scholar] [CrossRef]
- Baoune, H.; Aparicio, J.D.; Pucci, G.; Ould El Hadj-Khelil, A.; Polti, M.A. Bioremediation of petroleum-contaminated soils using Streptomyces sp. Hlh1. J. Soils Sediments 2019, 19, 2222–2230. [Google Scholar] [CrossRef]
- Boudh, S.; Tiwari, S.; Singh, J.S. Microbial-Mediated Lindane Bioremediation. In Agro-Environmental Sustainability: Volume 2: Managing Environmental Pollution; Singh, J.S., Seneviratne, G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 213–233. ISBN 978-3-319-49727-3. [Google Scholar]
- Medina, R.; Gara, P.M.D.; Fernández-González, A.J.; Rosso, J.A.; Del Panno, M.T. Remediation of a soil chronically contaminated with hydrocarbons through persulfate oxidation and bioremediation. Sci. Total Environ. 2018, 618, 518–530. [Google Scholar] [CrossRef]
- de Santos, A.A.; da Silveira, J.A.G.; Bonifacio, A.; Rodrigues, A.C.; do Figueiredo, M.V.B. Antioxidant response of cowpea co-inoculated with plant growth-promoting bacteria under salt stress. Brazilian J. Microbiol. Public. Brazilian Soc. Microbiol. 2018, 49, 513–521. [Google Scholar] [CrossRef]
- Jaemsaeng, R.; Jantasuriyarat, C.; Thamchaipenet, A. Molecular interaction of 1-aminocyclopropane-1-carboxylate deaminase (ACCD)-producing endophytic Streptomyces sp. GMKU 336 towards salt-stress resistance of Oryza sativa L. cv. KDML105. Sci. Rep. 2018, 8, 1950. [Google Scholar] [CrossRef]
- Tolba, S.T.M.; Ibrahim, M.; Amer, E.A.M.; Ahmed, D.A.M. First insights into salt tolerance improvement of Stevia by plant growth-promoting Streptomyces species. Arch. Microbiol. 2019, 201, 1295–1306. [Google Scholar] [CrossRef]
- Xu, L.; Naylor, D.; Dong, Z.; Simmons, T.; Pierroz, G.; Hixson, K.K.; Kim, Y.-M.; Zink, E.M.; Engbrecht, K.M.; Wang, Y.; et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E4284–E4293. [Google Scholar] [CrossRef] [Green Version]
- United States Environmental Protection Agency. Bioremediation. Contam. Site Clean-up Inf. 2020. Available online: https://clu-in.org/greenremediat (accessed on 9 February 2020).
- Vidali, M. Bioremediation. An overview*. Pure Appl. Chem. 2001, 73, 1163–1172. [Google Scholar] [CrossRef]
- Abatenh, E.; Gizaw, B.; Tsegaye, Z.; Wassie, M. The Role of Microorganisms in Bioremediation—A Review. Open J. Environ. Biol. 2017, 2, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, M.S.; Benimeli, C.S.; Cuozzo, S.A.; Amoroso, M.J. Isolation of pesticide-degrading actinomycetes from a contaminated site: Bacterial growth, removal and dechlorination of organochlorine pesticides. Int. Biodeterior. Biodegrad. 2010, 64, 434–441. [Google Scholar] [CrossRef]
- Fuentes, M.S.; Sáez, J.M.; Benimeli, C.S.; Amoroso, M.J. Lindane biodegradation by defined consortia of indigenous Streptomyces strains. Water Air Soil Pollut. 2011, 222, 217–231. [Google Scholar] [CrossRef]
- Alvarez, A.; Saez, J.M.; Davila Costa, J.S.; Colin, V.L.; Fuentes, M.S.; Cuozzo, S.A.; Benimeli, C.S.; Polti, M.A.; Amoroso, M.J. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef]
- Fuentes, M.S.; Briceño, G.E.; Saez, J.M.; Benimeli, C.S.; Diez, M.C.; Amoroso, M.J. Enhanced removal of a pesticides mixture by single cultures and consortia of free and immobilized Streptomyces strains. Biomed Res. Int. 2013, 2013, 9. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, M.S.; Alvarez, A.; Saez, J.M.; Benimeli, C.S.; Amoroso, M.J. Methoxychlor bioremediation by defined consortium of environmental Streptomyces strains. Int. J. Environ. Sci. Technol. 2014, 11, 1147–1156. [Google Scholar] [CrossRef]
- Benimeli, C.S.; Fuentes, M.S.; Abate, C.M.; Amoroso, M.J. Bioremediation of lindane-contaminated soil by Streptomyces sp. M7 and its effects on Zea mays growth. Int. Biodeterior. Biodegrad. 2007, 61, 233–239. [Google Scholar] [CrossRef]
- Cuozzo, S.A.; Fuentes, M.S.; Bourguignon, N.; Benimeli, C.S.; Amoroso, M.J. Chlordane biodegradation under aerobic conditions by indigenous Streptomyces strains. Int. Biodeterior. Biodegrad. 2012, 66, 19–24. [Google Scholar] [CrossRef]
- Schippers, A. Microorganisms involved in bioleaching and nucleic acid-based molecular methods for their identification and quantification. In Microbial Processing of Metal Sulfides; Donati, E.R., Sand, W., Eds.; Springer Netherlands: Dordrecht, The Netherland, 2007; pp. 3–33. ISBN 978-1-4020-5589-8. [Google Scholar]
- Polti, M.A.; Atjián, M.C.; Amoroso, M.J.; Abate, C.M. Soil chromium bioremediation: Synergic activity of actinobacteria and plants. Int. Biodeterior. Biodegrad. 2011, 65, 1175–1181. [Google Scholar] [CrossRef]
- Polti, M.A.; Amoroso, M.J.; Abate, C.M. Intracellular chromium accumulation by Streptomyces sp. MC1. Water Air Soil Pollut. 2011, 214, 49–57. [Google Scholar] [CrossRef]
- Albarracín, V.; Winik, B.; Kothe, E.; Amoroso, M.J.; Abate, J.C. Copper bioaccumulation by actinobacteria Amycolatopsis sp. AB0. J. Basic Microbiol. 2008, 48, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Polti, M.; Amoroso, M.J.; Abate, C.M. Chromium (VI) resistance and removal by actinomycete strains isolated from sediments. Chemosphere 2008, 67, 660–667. [Google Scholar] [CrossRef]
- Chen, M.; Xu, P.; Zeng, G.; Yang, C.; Huang, D.; Zhang, J. Bioremediation of soils contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting: Applications, microbes and future research needs. Biotechnol. Adv. 2015, 33, 745–755. [Google Scholar] [CrossRef]
- Gupta, A.; Joia, J.; Sood, A.; Sood, R.; Sidhu, C.; Kaur, G. Microbes as potential tool for remediation of heavy metals: A review. J. Microb. Biochem. Technol. 2016, 8, 364–372. [Google Scholar] [CrossRef] [Green Version]
- Moraga, N.B.; Poma, H.R.; Amoroso, M.J.; Rajal, V.B. Isolation and characterization of indigenous Streptomyces and Lentzea strains from soils containing boron compounds in Argentina. J. Basic Microbiol. 2014, 54, 568–577. [Google Scholar] [CrossRef]
- Moraga, N.B.; Amoroso, M.J.; Rajal, V.B. Streptomyces from soils contaminated with boron compounds. In Actinobacteria. Application in Bioremediation and Production of Industrial Enzymes; Amoroso, M.J., Benimeli, C.S., Cuozzo, S.A., Eds.; CRC Press: Boca Raton, FL, USA, 2013; pp. 136–164. [Google Scholar]
- Majzlik, P.; Strasky, A.; Adam, V.; Nemec, M.; Trnkova, L.; Zehnalek, J.; Hubalek, J.; Provaznik, I.; Kizek, R. Influence of zinc(II) and copper(II) ions on Streptomyces bacteria revealed by electrochemistry. Int. J. Electrochem. Sci. 2011, 6, 2171–2191. [Google Scholar]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef]
- Thavamani, P.; Samkumar, R.A.; Satheesh, V.; Subashchandrabose, S.R.; Ramadass, K.; Naidu, R.; Venkateswarlu, K.; Megharaj, M. Microbes from mined sites: Harnessing their potential for reclamation of derelict mine sites. Environ. Pollut. 2017, 230, 495–505. [Google Scholar] [CrossRef]
- Schütze, E.; Weist, A.; Klose, M.; Wach, T.; Schumann, M.; Nietzsche, S.; Merten, D.; Baumert, J.; Majzlan, J.; Kothe, E. Taking nature into lab: Biomineralization by heavy metal-resistant streptomycetes in soil. Biogeosciences 2013, 10, 3605–3614. [Google Scholar] [CrossRef] [Green Version]
- Pérez-González, T.; Valverde-Tercedor, C.; Jiménez-López, C. Biomineralización bacteriana de magnetita y aplicaciones. Semin. SEM 2012, 7, 58–74. [Google Scholar]
- Handley-Sidhu, S.; Renshaw, J.C.; Moriyama, S.; Stolpe, B.; Mennan, C.; Bagheriasl, S.; Yong, P.; Stamboulis, A.; Paterson-Beedle, M.; Sasaki, K.; et al. Uptake of Sr2+ and Co2+ into biogenic hydroxyapatite: Implications for biomineral ion exchange synthesis. Environ. Sci. Technol. 2011, 45, 6985–6990. [Google Scholar] [CrossRef] [PubMed]
- Lowenstam, H.A. Minerals formed by organisms. Science 1981, 211, 1126–1131. [Google Scholar] [CrossRef] [Green Version]
- Pal, A.; Paul, A. Microbial extracellular polymeric substances: Central elements in heavy metal bioremediation. Indian J. Microbiol. 2008, 48, 49–64. [Google Scholar] [CrossRef] [Green Version]
- Martínez, F.L.; Orce, I.G.; Rajal, V.B.; Irazusta, V.P. Salar del Hombre Muerto, source of lithium-tolerant bacteria. Environ. Geochem. Health 2018, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Moritz, R.; Kirschner, C.; Borchard, W.; Maibaum, R.; Wingender, J.; Flemming, H. The role of intermolecular interactions: Studies on model systems for bacterial biofilms. Int. Biodeterior. Biodegrad. 1999, 26, 3–16. [Google Scholar] [CrossRef]
- Centro de Información Minera de Argentina. Boratos. Panor. Merc. Rocas Miner. Ind. 2018. Available online: http://bit.ly/2SDslO9 (accessed on 9 February 2020).
- Zahran, H.H. Diversity, adaptation and activity of the bacterial flora in saline environments. Biol. Fertil. Soils 1997, 25, 211–223. [Google Scholar] [CrossRef]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica (Cairo) 2012, 2012, 15. [Google Scholar] [CrossRef] [Green Version]
- Palaniyandi, S.A.; Damodharan, K.; Yang, S.H.; Suh, J.W. Streptomyces sp. strain PGPA39 alleviates salt stress and promotes growth of “Micro Tom” tomato plants. J. Appl. Microbiol. 2014, 117, 766–773. [Google Scholar] [CrossRef]
- Srivastava, S.; Patel, J.S.; Singh, H.B.; Sinha, A.; Sarma, B.K. Streptomyces rochei SM3 induces stress tolerance in chickpea against Sclerotinia sclerotiorum and NaCl. J. Phytopathol. 2015, 163, 583–592. [Google Scholar] [CrossRef]
- Hasegawa, S.; Meguro, A.; Nishimura, T.; Kunoh, H. Drought tolerance of tissue-cultured seedlings of mountain laurel (Kalmia latifolia L.) induced by an endophytic actinomycete I. Enhancement of osmotic pressure in leaf cells. Actinomycetologica 2004, 18, 43–47. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, S.; Meguro, A.; Toyoda, K.; Nishimura, T.; Kunoh, H. Drought tolerance of tissue-cultured seedlings of mountain laurel (Kalmia latifolia L.) induced by an endophytic actinomycete II. Acceleration of callose accumulation and lignification. Actinomycetologica 2005, 19, 13–17. [Google Scholar] [CrossRef] [Green Version]
- Yandigeri, M.S.; Meena, K.K.; Singh, D.; Malviya, N.; Singh, D.P.; Solanki, M.K.; Yadav, A.K.; Arora, D.K. Drought-tolerant endophytic actinobacteria promote growth of wheat (Triticum aestivum) under water stress conditions. Plant Growth Regul. 2012, 68, 411–420. [Google Scholar] [CrossRef]
- Selvakumar, G.; Bhatt, R.M.; Upreti, K.K.; Bindu, G.H.; Shweta, K. Citricoccus zhacaiensis B-4 (MTCC 12119) a novel osmotolerant plant growth promoting actinobacterium enhances onion (Allium cepa L.) seed germination under osmotic stress conditions. World J. Microbiol. Biotechnol. 2015, 31, 833–839. [Google Scholar] [CrossRef]
- Kaur, T.; Vasudev, A.; Sohal, S.K.; Manhas, R.K. Insecticidal and growth inhibitory potential of Streptomyces hydrogenans DH16 on major pest of India, Spodoptera litura (Fab.) (Lepidoptera: Noctuidae). BMC Microbiol. 2014, 14, 227. [Google Scholar] [CrossRef] [Green Version]
- Gurney, K.A.; Mantle, P.G. Biosynthesis of 1-N-methylalbonoursin by an endophytic Streptomyces sp. isolated from perennial ryegrass. J. Nat. Prod. 1993, 56, 1194–1198. [Google Scholar] [CrossRef]
- Castillo, U.F.; Strobel, G.A.; Ford, E.J.; Hess, W.M.; Porter, H.; Jensen, J.B.; Albert, H.; Robison, R.; Condron, M.A.M.; Teplow, D.B.; et al. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscansa. Microbiology 2002, 148, 2675–2685. [Google Scholar] [CrossRef] [Green Version]
- Vijayabharathi, R.; Kumari, B.R.; Sathya, A.; Srinivas, V.; Abhishek, R.; Sharma, H.C.; Gopalakrishnan, S. Biological activity of entomopathogenic actinomycetes against lepidopteran insects (Noctuidae: Lepidoptera). Can. J. Plant Sci. 2014, 94, 759–769. [Google Scholar] [CrossRef]
- Saeed, E.E.; Sham, A.; Salmin, Z.; Abdelmowla, Y.; Iratni, R.; El-Tarabily, K.; AbuQamar, S. Streptomyces globosus UAE1, a potential effective biocontrol agent for black scorch disease in date palm plantations. Front. Microbiol. 2017, 8, 1455. [Google Scholar] [CrossRef]
- Loria, R.; Kers, J.; Joshi, M. Evolution of plant pathogenicity in Streptomyces. Annu. Rev. Phytopathol. 2006, 44, 469–487. [Google Scholar] [CrossRef] [PubMed]
- Loria, R.; Bignell, D.R.D.; Moll, S.; Huguet-Tapia, J.C.; Joshi, M.V.; Johnson, E.G.; Seipke, R.F.; Gibson, D.M. Thaxtomin biosynthesis: The path to plant pathogenicity in the genus Streptomyces. Antonie Van Leeuwenhoek 2008, 94, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, J.; Díaz-Cruz, G.; Cheng, Z.; Bignell, D.R.D. Virulence mechanisms of plant-pathogenic Streptomyces species: An updated review. Microbiology 2019, 165, 1025–1040. [Google Scholar] [CrossRef] [PubMed]
- Bignell, D.R.D.; Fyans, J.K.; Cheng, Z. Phytotoxins produced by plant pathogenic Streptomyces species. J. Appl. Microbiol. 2014, 116, 223–235. [Google Scholar] [CrossRef] [PubMed]
- El-Tarabily, K.A.; Sivasithamparam, K. Non-streptomycete actinomycetes as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Soil Biol. Biochem. 2006, 38, 1505–1520. [Google Scholar] [CrossRef]
- Cohen, M.F.; Mazzola, M. Resident bacteria, nitric oxide emission and particle size modulate the effect of Brassica napus seed meal on disease incited by Rhizoctonia solani and Pythium spp. Plant Soil 2006, 286, 75–86. [Google Scholar] [CrossRef]
- Mahmoudi, E.; Tabatabaei, B.E.S.; Venturi, V. Virulence attenuation of Pectobacterium carotovorum using N-Acyl-homoserine lactone degrading bacteria isolated from potato rhizosphere. Plant Pathol. J. 2011, 27, 242–248. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Qiu, Z.; You, J.; Tan, H.; Zhou, S. Isolation and characterization of endophytic streptomycete antagonists of fusarium wilt pathogen from surface-sterilized banana roots. FEMS Microbiol. Lett. 2005, 247, 147–152. [Google Scholar] [CrossRef]
- Getha, K.; Vikineswary, S.; Wong, W.H.; Seki, T.; Ward, A.; Goodfellow, M. Evaluation of Streptomyces sp. strain g10 for suppression of Fusarium wilt and rhizosphere colonization in pot-grown banana plantlets. J. Ind. Microbiol. Biotechnol. 2005, 32, 24–32. [Google Scholar] [CrossRef]
- Taechowisan, T.; Peberdy, J.F.; Lumyong, S. Isolation of endophytic actinomycetes from selected plants and their antifungal activity. World J. Microbiol. Biotechnol. 2003, 19, 381–385. [Google Scholar] [CrossRef]
- Miyashita, K.; Fujii, T.; Sawada, Y. Molecular cloning and characterization of chitinase genes from Streptomyces lividans 66. Microbiology 1991, 137, 2065–2072. [Google Scholar] [CrossRef] [Green Version]
- Abd-Allah, E.F. Streptomyces plicatus as a model biocontrol agent. Folia Microbiol. (Praha) 2001, 46, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Joo, G.-J. Production of an anti-fungal substance for biological control of Phytophthora capsici causing phytophthora blight in red-peppers by Streptomyces halstedii. Biotechnol. Lett. 2005, 27, 201–205. [Google Scholar] [CrossRef]
- Mahadevan, B.; Crawford, D.L. Properties of the chitinase of the antifungal biocontrol agent Streptomyces lydicus WYEC108. Enzyme Microb. Technol. 1997, 20, 489–493. [Google Scholar] [CrossRef]
- Errakhi, R.; Bouteau, F.; Lebrihi, A.; Barakate, M. Evidences of biological control capacities of Streptomyces spp. against Sclerotium rolfsii responsible for damping-off disease in sugar beet (Beta vulgaris L.). World J. Microbiol. Biotechnol. 2007, 23, 1503–1509. [Google Scholar] [CrossRef]
- Trejo-Estrada, S.R.; Paszczynski, A.; Crawford, D.L. Antibiotics and enzymes produced by the biocontrol agent Streptomyces violaceusniger YCED-9. J. Ind. Microbiol. Biotechnol. 1998, 21, 81–90. [Google Scholar] [CrossRef]
- Barakate, M.; Ouhdouch, Y.; Oufdou, K.; Beaulieu, C. Characterization of rhizospheric soil streptomycetes from Moroccan habitats and their antimicrobial activities. World J. Microbiol. Biotechnol. 2002, 18, 49–54. [Google Scholar] [CrossRef]
- Emmert, E.A.; Handelsman, J. Biocontrol of plant disease: A (Gram-) positive perspective. FEMS Microbiol. Lett. 1999, 171, 1–9. [Google Scholar] [CrossRef]
- Vijayabharathi, R.; Gopalakrishnan, S.; Sathya, A.; Kumar, M.V.; Srinivas, V.; Mamta, S. Streptomyces sp. as plant growth-promoters and host-plant resistance inducers against Botrytis cinerea in chickpea. Biocontrol Sci. Technol. 2018, 28, 1140–1163. [Google Scholar] [CrossRef]
- Vatsa-Portugal, P.; Aziz, A.; Rondeau, M.; Villaume, S.; Morjani, H.; Clément, C.; Ait Barka, E. How Streptomyces anulatus primes grapevine defenses to cope with gray mold: A study of the early responses of cell suspensions. Front. Plant Sci. 2017, 8, 1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baz, M.; Tran, D.; Kettani-Halabi, M.; Samri, S.E.; Jamjari, A.; Biligui, B.; Meimoun, P.; El-Maarouf-Bouteau, H.; Garmier, M.; Saindrenan, P.; et al. Calcium- and ROS-mediated defence responses in BY2 tobacco cells by nonpathogenic Streptomyces sp. J. Appl. Microbiol. 2012, 112, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Poschenrieder, C.; Amenós, M.; Corrales, I.; Doncheva, S.; Barceló, J. Root behavior in response to aluminum toxicity. In Plant-Environment Interactions. Signaling and Communication in Plants; Baluška, F., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 21–43. ISBN 978-3-540-89230-4. [Google Scholar]
- Dahal, B.; NandaKafle, G.; Perkins, L.; Brözel, V.S. Diversity of free-Living nitrogen fixing Streptomyces in soils of the badlands of South Dakota. Microbiol. Res. 2017, 195, 31–39. [Google Scholar] [CrossRef] [PubMed]
- MacKellar, D.; Lieber, L.; Norman, J.S.; Bolger, A.; Tobin, C.; Murray, J.W.; Oksaksin, M.; Chang, R.L.; Ford, T.J.; Nguyen, P.Q.; et al. Streptomyces thermoautotrophicus does not fix nitrogen. Sci. Rep. 2016, 6, 20086. [Google Scholar] [CrossRef]
- Aly, M.M.; Tork, S.; Al-Garni, S.M.; Kabli, S.A. Production and characterization of phytase from Streptomyces luteogriseus R10 isolated from decaying wood samples. Int. J. Agric. Biol. 2015, 17, 515–522. [Google Scholar]
- Ghorbani-Nasrabadi, R.; Greiner, R.; Alikhani, H.A.; Hamedi, J. Identification and determination of extracellular phytate-degrading activity in actinomycetes. World J. Microbiol. Biotechnol. 2012, 28, 2601–2608. [Google Scholar] [CrossRef]
- Farhat, M.B.; Boukhris, I.; Chouayekh, H. Mineral phosphate solubilization by Streptomyces sp. CTM396 involves the excretion of gluconic acid and is stimulated by humic acids. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [Green Version]
- Jog, R.; Pandya, M.; Nareshkumar, G.; Rajkumar, S. Mechanism of phosphate solubilization and antifungal activity of Streptomyces spp. isolated from wheat roots and rhizosphere and their application in improving plant growth. Microbiology 2014, 160, 778–788. [Google Scholar] [CrossRef] [Green Version]
- Rashad, F.M.; Fathy, H.M.; El-Zayat, A.S.; Elghonaimy, A.M. Isolation and characterization of multifunctional Streptomyces species with antimicrobial, nematicidal and phytohormone activities from marine environments in Egypt. Microbiol. Res. 2015, 175, 34–47. [Google Scholar] [CrossRef]
- Waraich, E.A.; Ahmad, R.; Halim, A.; Aziz, T. Alleviation of temperature stress by nutrient management in crop plants: A review. J. Soil Sci. Plant Nutr. 2012, 12, 221–244. [Google Scholar] [CrossRef] [Green Version]
- Root, T.L.; Price, J.T.; Hall, K.R.; Schneider, S.H.; Rosenzweig, C.; Pounds, J.A. Fingerprints of global warming on wild animals and plants. Nature 2003, 421, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Jaemsaeng, R.; Jantasuriyarat, C.; Thamchaipenet, A. Positive role of 1-aminocyclopropane-1-carboxylate deaminase-producing endophytic Streptomyces sp. GMKU 336 on flooding resistance of mung bean. Agric. Nat. Resour. 2018, 52, 330–334. [Google Scholar] [CrossRef]
- Doaa, D.B.; El-Saeed, S.A. Assessment of the optimum conditions for production and purification of superoxide dismutase from Actinomycetes. Egypt. J. Exp. Biol. 2016, 12, 163–173. [Google Scholar]
- Tan, L.T.-H.; Chan, K.-G.; Khan, T.M.; Bukhari, S.I.; Saokaew, S.; Duangjai, A.; Pusparajah, P.; Lee, L.-H.; Goh, B.-H. Streptomyces sp. MUM212 as a source of antioxidants with radical scavenging and metal chelating properties. Front. Pharmacol. 2017, 8, 276. [Google Scholar] [CrossRef]
- Wang, Z.; Solanki, M.K.; Yu, Z.-X.; Yang, L.-T.; An, Q.-L.; Dong, D.-F.; Li, Y.-R. Draft genome analysis offers insights into the mechanism by which Streptomyces chartreusis WZS021 increases drought tolerance in sugarcane. Front. Microbiol. 2019, 9, 3262. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef] [Green Version]
- Chhabra, R. Soil Salinity and Water Quality; A A Balkema Publishers: Rotterdam, The Netherlands, 1996; ISBN 9789054107279. [Google Scholar]
- FAO (Food and Agriculture Organization of the United Nations). Handbook for Saline Soil Management; Vargas, R., Pankova, E.I., Balyuk, S.A., Krasilnikov, P.V., Khasankhanova, G., Eds.; Food and Agriculture Organization of the United Nations and Lomonosov Moscow State University: Rome, Italy, 2018; ISBN 9789251301418. [Google Scholar]
- Taleisnik, E.; Lavado, R. Ambientes Salinos y Alcalinos de la Argentina: Recursos y Aprovechamiento Productivo; EUDEBA: Buenos Aires, Argentina, 2016. [Google Scholar]
- Sparks, D. The Chemistry of Saline and Sodic soils. In Environmental Soil Chemistry; Academic Press Inc.: San Diego, CA, USA, 2003; pp. 285–300. ISBN 9780126564464. [Google Scholar]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Läuchli, A.; Grattan, S.R. Plant Responses to Saline and Sodic Conditions. In Agricultural Salinity Assessment and Management; Wallender, W.W., Tanji, K.K., Eds.; ASCE: Reston, VA, USA, 2011; pp. 169–205. ISBN 9780784411698. [Google Scholar]
- Machado, R.M.A.; Serralheiro, R.P. Soil salinity: Effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- López-Berenguer, C.; García-Viguera, C.; Carvajal, M. Are root hydraulic conductivity responses to salinity controlled by aquaporins in broccoli plants? Plant Soil 2006, 279, 13–23. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [Green Version]
- Ors, S.; Suarez, D.L. Spinach biomass yield and physiological response to interactive salinity and water stress. Agric. Water Manag. 2017, 190, 31–41. [Google Scholar] [CrossRef]
- Giuffrida, F.; Scuderi, D.; Giurato, R.; Leonardi, C. Physiological response of broccoli and cauliflower as affected by NaCl salinity. Acta Hortic. 2013, 1005, 435–442. [Google Scholar] [CrossRef]
- Butcher, K.; Wick, A.F.; Desutter, T.; Chatterjee, A.; Harmon, J. Soil salinity: A threat to global food security. Agron. J. 2016, 108, 2189–2200. [Google Scholar] [CrossRef]
- Khan, M.S.A. Effects of salt and water stress on leaf production, sodium and potassium ion accumulation in soybean. J. Plant Sci. (Science Publ. Group) 2014, 2, 209. [Google Scholar] [CrossRef] [Green Version]
- Queiroz, H.M.; Sodek, L.; Haddad, C.R.B. Effect of salt on the growth and metabolism of Glycine max. Brazilian Arch. Biol. Technol. 2012, 55, 809–817. [Google Scholar] [CrossRef] [Green Version]
- Ullah, A.; Li, M.; Noor, J.; Tariq, A.; Liu, Y.; Shi, L. Effects of salinity on photosynthetic traits, ion homeostasis and nitrogen metabolism in wild and cultivated soybean. PeerJ 2019, 7, e8191. [Google Scholar] [CrossRef] [Green Version]
- Sumner, M.E.; Naidu, R. Sodic Soils: Distribution, Properties, Management and Environmental Consequences; Oxford University Press Inc.: New York, NY, USA, 1998; ISBN 9780195096552. [Google Scholar]
- Gambrell, R.; Wiesepape, J.B.; Patrick, W.H., Jr.; Duff, M.C. The effects of pH, redox, and salinity on metal release from a contaminated sediment. Water Air Soil Pollut. 1991, 57–58, 359–367. [Google Scholar] [CrossRef]
- McLaughlin, M.J.; Tiller, K.G. Chloro-complexation of cadmium in soil solutions of saline-sodic soils increases phyto-availability of cadmium. Trans. 15th World Congr. Soil Sci. 1994, 3b, 195–196. [Google Scholar]
- Wicke, B.; Smeets, E.; Dornburg, V.; Vashev, B.; Gaiser, T.; Turkenburg, W.; Faaij, A. The global technical and economic potential of bioenergy from salt-affected soils. Energy Environ. Sci. 2011, 4, 2669–2681. [Google Scholar] [CrossRef] [Green Version]
- Richards, L. Diagnosis and Improvement of Saline Alkali Soils; Laboratory, U.S., Ed.; Government Printing Office: Washington, DC, USA, 1954. [Google Scholar]
- Keren, R.; Myamoto, S. Reclamation of saline, sodic and boron-affected soils. Agric. Salin. Assess. Manag. 1990, 71, 410–431. [Google Scholar]
- Zaman, M.; Shahid, S.A.; Heng, L. Soil Salinity: Historical Perspectives and a World Overview of the Problem. In Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Springer International Publishing AG: Cham, Switzerland, 2018; p. 17. ISBN 9783319961903. [Google Scholar]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- FAO/IIASA/ISRIC/ISS-CAS/JRC. Harmonized World Soil Database (version 1.1); Food and Agriculture Organization of the United Nations, FAO: Rome, Italy; IIASA: Laxenburg, Austria, 2009. [Google Scholar]
- FAO (Food and Agriculture Organization of the United Nations). Advances in the Assessment and Mmonitoring of Salinization and Status of Biosaline Agriculture; FAO: Rome, Italy, 2010; ISBN 9789251064399. [Google Scholar]
- FAO (Food and Agriculture Organization of the United Nations). FAOSTAT. Stat. Div. UN Food Agric. Organ. 2016. Available online: http://www.fao.org/faostat/en/ (accessed on 14 December 2019).
- Ladeiro, B. Saline agriculture in the 21st century: Using salt contaminated resources to cope food requirements. J. Bot. 2012, 2012, 310705. [Google Scholar] [CrossRef]
- Keren, R. Salt-affected Soils, Reclamation. In Encyclopedia of Soils in the Environment; Hillel, D., Ed.; Academic Press Inc.: San Diego, CA, USA, 2005; pp. 454–461. ISBN 9780123485304. [Google Scholar]
- Bradshaw, A.D. Ecological principles and land reclamation practice. Landsc. Plan. 1984, 11, 35–48. [Google Scholar] [CrossRef]
- Gupta, R.K.; Abrol, I.P. Salt-Affected Soils: Their Reclamation and Management for Crop Production. In Advances in Soil Science; Lal, R., Ed.; Springer: New York, NY, USA, 1990; pp. 224–261. [Google Scholar]
- Pollmann, K.; Kutschke, S.; Matys, S.; Raff, J.; Hlawacek, G.; Lederer, F.L. Bio-recycling of metals: Recycling of technical products using biological applications. Biotechnol. Adv. 2018, 36, 1048–1062. [Google Scholar] [CrossRef]
- Arora, S.; Vanza, M.J.; Mehta, R.; Bhuva, C.; Patel, P.N. Halophilic microbes for bio-remediation of salt affected soils. African J. Microbiol. Res. 2014, 8, 3070–3078. [Google Scholar]
- Grover, M.; Ali, S.Z.; Sandhya, V.; Rasul, A.; Venkateswarlu, B. Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J. Microbiol. Biotechnol. 2011, 27, 1231–1240. [Google Scholar] [CrossRef]
- Ashraf, M.; Hasnain, S.; Berge, O.; Mahmood, T. Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol. Fertil. Soils 2004, 40, 157–162. [Google Scholar] [CrossRef]
- Roberson, E.B.; Firestone, M.K. Relationship between desiccation and exopolysaccharide production in a soil Pseudomonas sp. Appl. Environ. Microbiol. 1992, 58, 1284–1291. [Google Scholar] [CrossRef] [Green Version]
- Tisdall, J.M.; Oades, J.M. Organic matter and water-stable aggregates in soils. Eur. J. Soil Sci. 1982, 33, 141–163. [Google Scholar] [CrossRef]
- Jeschke, W.D.; Wolf, O. Effect of NaCI salinity on growth, development, ion distribution, and ion translocation in castor bean (Ricinus communis L.). J. Plant Physiol. 1988, 132, 45–53. [Google Scholar] [CrossRef]
- Oren, A. Industrial and environmental applications of halophilic microorganisms. Environ. Technol. 2010, 31, 825–834. [Google Scholar] [CrossRef] [Green Version]
- Sandhya, V.; Ali, Z.; Grover, M.; Reddy, G.; Venkateswarlu, B. Alleviation of drought stress effects in sunflower seedlings by the exopolysaccharides producing Pseudomonas putida strain GAP-p45. Biol. Fertil. Soils 2009, 46, 17–26. [Google Scholar] [CrossRef]
- Arora, S.; Vanza, M. Microbial Approach for Bioremediation of Saline and Sodic Soils. In Bioremediation of Salt Affected Soils: An Indian Perspective; Arora, S., Singh, A.K., Singh, Y.P., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 87–100. ISBN 9783319482569. [Google Scholar]
- Ventosa, A.; Márquez, M.C.; Garabito, M.J.; Arahal, D.R. Moderately halophilic gram-positive bacterial diversity in hypersaline environments. Extremophiles 1998, 2, 297–304. [Google Scholar] [CrossRef]
- Oren, A. Diversity of halophilic microorganisms : Environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 2002, 28, 56–63. [Google Scholar] [CrossRef]
- Vasavada, S.H.; Thumar, J.T.; Singh, S.P. Secretion of a potent antibiotic by salt-tolerant and alkaliphilic actinomycete Streptomyces sannanensis strain RJT-1. Curr. Sci. 2006, 91, 1393–1397. [Google Scholar]
- Kannabiran, K.; Deepika Lakshmipathy, T.; Arun Prasad, A.; Kannabiran, K. Production of biosurfactant and heavy metal resistance activity of Streptomyces Sp. VITDDK3-a novel halo tolerant actinomycetes isolated from Saltpan soil. Adv. Biol. Res. (Rennes) 2010, 4, 108–115. [Google Scholar]
- Margesin, M.R. Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl. Microbiol. Biotechnol. 2001, 56, 650–663. [Google Scholar] [CrossRef]
- Bursy, J.; Kuhlmann, A.U.; Pittelkow, M.; Hartmann, H.; Jebbar, M.; Pierik, A.J.; Bremer, E. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol. 2008, 74, 7286–7296. [Google Scholar] [CrossRef] [Green Version]
- Empadinhas, N.; Da Costa, M.S. Osmoadaptation mechanisms in prokaryotes: Distribution of compatible solutes. Int. Microbiol. 2008, 11, 151–161. [Google Scholar]
- Killham, K.; Firestone, M.K. Salt stress control of intracellular solutes in streptomycetes indigenous to saline soils. Appl. Environ. Microbiol. 1984, 47, 301–306. [Google Scholar] [CrossRef] [Green Version]
- Kol, S.; Elena Merlo, M.; Scheltema, R.A.; De Vries, M.; Vonk, R.J.; Kikkert, N.A.; Dijkhuizen, L.; Breitling, R.; Takano, E. Metabolomic characterization of the salt stress response in Streptomyces coelicolor. Appl. Environ. Microbiol. 2010, 76, 2574–2581. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, K.; Takahashi, Y.; Berberich, T.; Imai, A.; Miyazaki, A.; Takahashi, T.; Michael, A.; Kusano, T. The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett. 2006, 580, 6783–6788. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, A.; Soltani, B.M.; Jouzani, G.S.; Karimi, E.; Nekouei, M.K.; Sadeghizadeh, M. Taxonomic study of a salt tolerant Streptomyces sp. strain C-2012 and the effect of salt and ectoine on lon expression level. Microbiol. Res. 2014, 169, 232–238. [Google Scholar] [CrossRef]
- Sobczyk, A.; Bellier, A.; Viala, J.; Mazodier, P. The Ion gene, encoding an ATP-dependent protease, is a novel member of the HAIR/HspR stress-response regulon in actinomycetes. Microbiology 2002, 148, 1931–1937. [Google Scholar] [CrossRef] [Green Version]
- Thumar, J.T.; Singh, S.P. Organic solvent tolerance of an alkaline protease from salt-tolerant alkaliphilic Streptomyces clavuligerus strain Mit-1. J. Ind. Microbiol. Biotechnol. 2009, 36, 211–218. [Google Scholar] [CrossRef]
- Vicente, M.F.; Gorrochategui, J.; Cabello, A.; Gonza, A.; Genilloud, O.; Basilio, A.; Gonza, I. Patterns of antimicrobial activities from soil actinomycetes isolated under different conditions of pH and salinity. J. Appl. Microbiol. 2003, 95, 814–823. [Google Scholar]
- Sajid, I.; Blandine, Æ.C.; Fondja, F. Antifungal and antibacterial activities of indigenous Streptomyces isolates from saline farmlands : Prescreening, ribotyping and metabolic diversity. World J. Microbiol. Biotechnol. 2009, 25, 601–610. [Google Scholar] [CrossRef]
- Palaniyandi, S.A.; Yang, S.H.; Zhang, L.; Suh, J.W. Effects of actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 2013, 97, 9621–9636. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Pande, S.; Sharma, M.; Humayun, P.; Kiran, B.K.; Sandeep, D.; Vidya, M.S.; Deepthi, K.; Rupela, O. Evaluation of actinomycete isolates obtained from herbal vermicompost for the biological control of Fusarium wilt of chickpea. Crop Prot. 2011, 30, 1070–1078. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Kloepper, J.W.; Ryu, C. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2008, 14, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Feng, Y. Characterization of nitrogen-fixing moderate halophilic cyanobacteria isolated from saline soils of Songnen Plain in China. Prog. Natural Sci. 2008, 18, 769–773. [Google Scholar] [CrossRef]
- Colwell, R.R. Microbial diversity: The importance of exploration and conservation. J. Ind. Microbiol. Biotechnol. 1997, 18, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Kloepper, J.W. Host specificity in microbe-microbe interactions: Biological control agents vary in specificity for hosts, pathogen control, ecological habitat, and environmental conditions. Bioscience 1996, 46, 406–409. [Google Scholar] [CrossRef] [Green Version]
- Gardner, J.M.; Chandler, J.L.; Feldman, A.W. Growth promotion and inhibition by antibiotic-producing fluorescent pseudomonads on citrus roots. Plant Soil 1984, 77, 103–113. [Google Scholar] [CrossRef]
- O’Neill, G.A.; Radley, R.A.; Chanway, C.P. Variable effects of emergence-promoting rhizobacteria on conifer seedling growth under nursery conditions. Biol. Fertil. Soils 1992, 13, 45–49. [Google Scholar] [CrossRef]
- Garcia, C.; Hernandez, T. Influence of salinity on the biological and biochemical activity of a calciorthird soil. Plant Soil 1996, 178, 255–263. [Google Scholar] [CrossRef]
- Zhao, B.; Wang, H.; Mao, X.; Li, R. Biodegradation of phenanthrene by a halophilic bacterial consortium under aerobic conditions. Curr. Microbiol. 2009, 58, 205–210. [Google Scholar] [CrossRef]
- Amoozegar, M.A.; Hamedi, J.; Dadashipour, M.; Shariatpanahi, S. Effect of salinity on the tolerance to toxic metals and oxyanions in native moderately halophilic spore-forming bacilli. World J. Microbiol. Biotechnol. 2005, 21, 1237–1243. [Google Scholar] [CrossRef]
- Betancur-Galvis, L.A.; Alvarez-Bernal, D.; Ramos-Valdivia, A.C.; Dendooven, L. Bioremediation of polycyclic aromatic hydrocarbon-contaminated saline-alkaline soils of the former Lake Texcoco. Chemosphere 2006, 62, 1749–1760. [Google Scholar] [CrossRef]
- Ngwenya, B.T. Bacterial Mineralization. In Reference Module in Materials Science and Materials Engineering; Elsevier: Amsterdam, The Netherlands, 2016; ISBN 9780128035818. Available online: http://bit.ly/2OGxVxJ (accessed on 14 December 2019).
- Theng, B.K.; Yuan, G. Nanoparticles in the soil environment. Elements 2008, 4, 395–399. [Google Scholar] [CrossRef]
- Prozorov, T.; Bazylinski, D.A.; Mallapragada, S.K.; Prozorov, R. Novel magnetic nanomaterials inspired by magnetotactic bacteria: Topical review. Mater. Sci. Eng. R Reports 2013, 74, 133–172. [Google Scholar] [CrossRef]
- Hennebel, T.; De Gusseme, B.; Boon, N.; Verstraete, W. Biogenic metals in advanced water treatment. Trends Biotechnol. 2009, 27, 90–98. [Google Scholar] [CrossRef]






| Potential | Species | Characteristics/Purposes |
|---|---|---|
| Bioremediation | ||
| Streptomyces albogriseolus 053 HQ538724.1 and S. lincolnensis 128 HQ538726.1 | Formation of boron minerals by the cells [6] | |
| Streptomyces sp. DPUA1566 | Production of a new biosurfactant lipoprotein for use in agro-industrial waste [53] | |
| Streptomyces sp. Hlh1 | Degradation of petroleum compounds in contaminated soils [54] | |
| Streptomyces sp. strain M7 | Possible lindane degradation [55] | |
| Streptomyces antioxidans MUSC164T | Remediation of soils chronically contaminated with hydrocarbons [56] | |
| Plant Growth Promotion | ||
| Streptomyces T5 | Increase of superoxide dismutase, catalase and phenol peroxidase activities in nodules of cowpea plants exposed to salt stress [57] | |
| Streptomyces sp. GMKU 336 | Increase of salt-stress resistance of Oryza sativa L. cv. KDML105 [58] | |
| Streptomyces spp. | Increase of salt tolerance of Stevia [59] | |
| Streptomyces coelicolor (Sc1) and Streptomyces ambofaciens (Sc2) | Colonization of roots during drought to improve plant growth [60] | |
| Region | Population (Millions) | Land Area with Irrigation a (thousand ha) | Arable Land b (thousand ha) | Permanent Crops c (thousand ha) | Salt-Affected Land d (thousand ha) |
|---|---|---|---|---|---|
| World | 7043 | 324,548 | 1,395,490 | 162,100 | 971 |
| Africa | 1077 | 15,265 | 230,862 | 33,571 | 295 |
| North America | 346 | 27,730 | 194,640 | 7526 | 63 |
| Central America | 163 | 7306 | 28,195 | 5178 | 4 |
| South America | 400 | 15,880 | 133,326 | 14,199 | 57 |
| Asia | 4240 | 228,667 | 480,140 | 83,495 | 291 |
| Europe | 738 | 25,414 | 274,151 | 15,211 | 2 |
| Oceania | 37 | 3261 | 48,702 | 1603 | 144 |
| Former USSR | 117 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romano-Armada, N.; Yañez-Yazlle, M.F.; Irazusta, V.P.; Rajal, V.B.; Moraga, N.B. Potential of Bioremediation and PGP Traits in Streptomyces as Strategies for Bio-Reclamation of Salt-Affected Soils for Agriculture. Pathogens 2020, 9, 117. https://doi.org/10.3390/pathogens9020117
Romano-Armada N, Yañez-Yazlle MF, Irazusta VP, Rajal VB, Moraga NB. Potential of Bioremediation and PGP Traits in Streptomyces as Strategies for Bio-Reclamation of Salt-Affected Soils for Agriculture. Pathogens. 2020; 9(2):117. https://doi.org/10.3390/pathogens9020117
Chicago/Turabian StyleRomano-Armada, Neli, María Florencia Yañez-Yazlle, Verónica P. Irazusta, Verónica B. Rajal, and Norma B. Moraga. 2020. "Potential of Bioremediation and PGP Traits in Streptomyces as Strategies for Bio-Reclamation of Salt-Affected Soils for Agriculture" Pathogens 9, no. 2: 117. https://doi.org/10.3390/pathogens9020117
APA StyleRomano-Armada, N., Yañez-Yazlle, M. F., Irazusta, V. P., Rajal, V. B., & Moraga, N. B. (2020). Potential of Bioremediation and PGP Traits in Streptomyces as Strategies for Bio-Reclamation of Salt-Affected Soils for Agriculture. Pathogens, 9(2), 117. https://doi.org/10.3390/pathogens9020117





