Validation of a Laboratory-Developed Triplex Molecular Assay for Simultaneous Detection of Gastrointestinal Adenovirus and Rotavirus in Stool Specimens
Abstract
1. Introduction
2. Materials and Methods
2.1. Testing Methods
2.2. RNA and DNA Extraction
2.3. PCR Amplification
2.4. Accuracy
2.5. Analytical Sensitivity: Limit of Detection
2.6. Precision: Repeatability (Intra Assay Precision) and Reproducibility (Inter Assay Precision)
2.7. Turnaround Time
2.8. Analytical Specificity
2.9. Ethics Statement
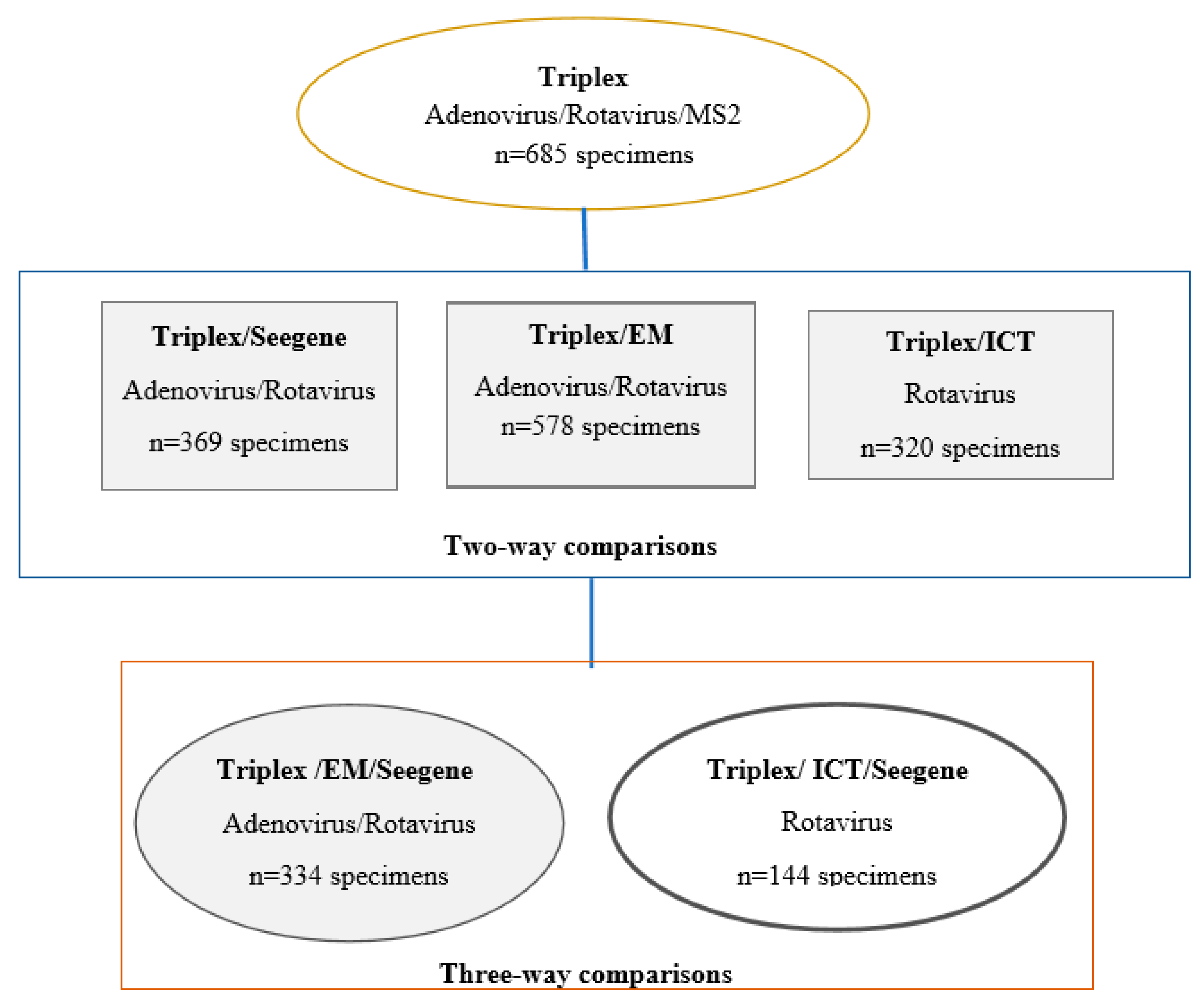
3. Results
4. Accuracy
4.1. Performance Characteristics of Triplex Assay Compared to Seegene PCR Assay
4.2. Performance Characteristics of Triplex Assay Against EM or Rotavirus ICT (Two Non-Standard Reference Testing Methods)
4.3. Three-Way Comparison of Triplex Assay Against EM and/or ICT
4.4. Adenovirus (Triplex-EM-Seegene)
4.5. Rotavirus (Triplex-EM-Seegene)
4.6. Rotavirus (Triplex-ICT-Seegene)
4.7. Analytical Sensitivity: Limit of Detection
4.8. Analytical Specificity
4.9. Interference
4.10. Precision and Turnaround Time
5. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lopman, B.A.; Reacher, M.H.; Van Duijnhoven, Y.; Hanon, F.X.; Brown, D.; Koopmans, M. Viral gastroenteritis outbreaks in Europe, 1995–2000. Emerg. Infect. Dis. 2003, 9, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, M. Outbreaks of viral gastroenteritis: What’s new in 2004? Curr. Opin. Infect. Dis. 2005, 18, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Krones, E.; Hogenauer, C. Diarrhea in the immunocompromised patient. Gastroenterol. Clin. North. Am. 2012, 41, 677–701. [Google Scholar] [CrossRef] [PubMed]
- Blanton, L.H.; Adams, S.M.; Berard, R.S.; Wei, G.; Bulens, S.N.; Widdowson, M.A.; Glass, R.I.; Monroe, S.S. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000–2004. J. Infect. Dis. 2006, 193, 413–421. [Google Scholar] [CrossRef]
- Svraka, S.; Duizer, E.; Venema, H.; de Bruin, E.; van der Veer, B.; Dorrestejn, B.; Keoopmans, M. Etiological role of viruses in outbreaks of acute gastroenteritis in the Netherlands from 1994 through 2005. J. Clin. Microbiol. 2007, 45, 1389–1394. [Google Scholar] [CrossRef]
- Isabel, S.; Higgins, R.R.; Peci, A.; Isabel, M.R.; Deeks, S.L.; Gubbay, J. Rotavirus genotypes circulating in Ontario, Canada, before and after implementation of the rotavirus immunization program. Vaccine 2018, 36, 2033–2040. [Google Scholar] [CrossRef]
- Girard, M.P.; Steele, D.; Chaignat, C.L.; Kieny, M.P. A review of vaccine research and development: Human enteric infections. Vaccine 2006, 24, 2732–2750. [Google Scholar] [CrossRef]
- Glass, R.I.; Noel, J.; Ando, T.; Fankhauser, R.; Belliot, G.; Mounts, A.; Parashar, U.D.; Breese, J.S.; Monroe, S.S. The epidemiology of enteric caliciviruses from humans: A reassessment using new diagnostics. J. Infect. Dis. 2000, 181, 254–261. [Google Scholar] [CrossRef]
- CDC. Rotavirus. Available online: https://www.cdc.gov/rotavirus/index.html (accessed on 4 June 2019).
- Pereira, L.A.; Ferreira, C.E.; Turchetto, G.D.; Nogueira, M.B.; Vidal, L.R.; Cruz, C.R.; Debur, M.C.; de Almeida, S.M.; Raboni, S.M. Molecular characterization of rotavirus genotypes in immunosuppressed and non-immunosuppressed pediatric patients. J. Pediatr. 2013, 89, 278–285. [Google Scholar] [CrossRef][Green Version]
- Wilson, S.E.; Rosella, L.C.; Wang, J.; Renaud, A.; Le Saux, N.; Crowcroft, N.S.; Desai, S.; Harris, T.; Bolotin, S.; Gubbay, J.; et al. Equity and impact: Ontario’s infant rotavirus immunization program five years following implementation. A population based cohort study. Vaccine 2019, 37, 2408–2414. [Google Scholar] [CrossRef]
- Uhnoo, I.; Wadell, G.; Svensson, L.; Johansson, M.E. Importance of enteric Adenoviruses 40 and 41 in acute gastroenteritis in infants and young children. J. Clin Microbiol. 1984, 20, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Baerjee, A.; De, P.; Manna, B.; Chawla-Sarkar, M. Molecular characterization of enteric adenovirus genotypes 40 and 41 identified in children with acute gastroenteritis in Kolkata, India during 2013–2014. J. Med. Virol. 2017, 89, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Matthes-Martin, S.; Boztug, H.; Lion, T. Diagnosis and treatment of adenovirus infection in immunocompromised patients. Expert Rev. Anti Infect. Ther. 2013, 11, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
- Lion, T.; Kosulin, K.; Landlinger, C.; Rauch, M.; Preuner, S.; Jugovic, D.; Pötschger, U.; Lawitschka, A.; Peters, C.; Fritsch, G.; et al. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia 2010, 24, 706–714. [Google Scholar] [CrossRef]
- Ibrahim, S.B.; El-Bialy, A.A.; Mohammed, M.S.; El-Sheikh, A.O.; Elhewala, A.; Bahgat, S. Detection of Rotavirus in children with acute gastroenteritis in Zagazig University Hospitals in Egypt. Electron. Physician. 2015, 7, 1227–1233. [Google Scholar] [CrossRef]
- Damen, M.; Minnaar, R.; Glasius, P.; vanderHam, A.; Koen, G.; Wertheim, P.; Beld, M. Real-time PCR with an internal control for detection of all known human adenovirus serotypes. J. Clin. Microbiol. 2008, 46, 3997–4003. [Google Scholar] [CrossRef][Green Version]
- Zeng, S.Q.; Halkosalo, A.; Salminen, M.; Szakal, E.D.; Puustinen, L.; Vesikari, T. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J. Virol. Meth. 2008, 153, 238–240. [Google Scholar] [CrossRef]
- Bennett, S.; Gunson, R. The development of a multiplex real-time RT-PCR for the detection of adenovirus, astrovirus, rotavirus and sapovirus from stool samples. J. Virol. Meth. 2017, 242, 30–34. [Google Scholar] [CrossRef]
- Higgins, R.R.; Beniprashad, M.; Cardona, M.; Masney, S.; Low, D.E.; Gubbay, J.B. Evaluation and Verification of the Seeplex Diarrhea-V ACE Assay for Simultaneous Detection of Adenovirus, Rotavirus and Norovirus Genogroups I and II in Clinical Stool Specimens. J. Clin. Microb. 2011, 49, 3154–3162. [Google Scholar] [CrossRef]
- Garcia, L.; Isenberg, H. Clinical Microbiology Procedures Handbook, 2nd ed.; ASM Press: Santa Monica, CA, USA, 2007. [Google Scholar]
- Barber, R.D.; Harmer, D.W.; Coleman, R.A.; Clark, B.J. GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genomics. 2005, 21, 389–395. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed]
- Layton, A.; McKay, L.; Williams, D.; Garret, V.; Gentry, R.; Sayler, G. Development of Bacteroides 16S rRNA Gene TaqMan-Based Real-Time PCR Assays for Estimation of Total, Human, and Bovine Fecal Pollution in Water. Appl. Environ. Microbiol. 2006, 72, 4214–4224. [Google Scholar] [CrossRef] [PubMed]
- Dreier, J.; Stomer, M.; Kleesiek, K. Use of bacteriphage MS2 as an internal control in viral reverse transcription-PCR assays. J. Clin. Mirobiol. 2005, 43, 4551–4557. [Google Scholar] [CrossRef] [PubMed]
- FDA. Guidance for Industry and FDA Staff. Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests. 2007. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/statistical-guidance-reporting-results-studies-evaluating-diagnostic-tests-guidance-industry-and-fda (accessed on 19 September 2018).
- Soltan, M.A.; Tsai, Y.L.; Lee, P.A.; Tsai, C.F.; Chang, H.G.; Wang, H.T.; Wilkes, R.P. Comparison of electron microscopy, ELISA, real time RT-PCR and insulated isothermal RT-PCR for the detection of Rotavirus group A (RVA) in feces of different animal species. J. Vir. Meth. 2016, 235, 99–104. [Google Scholar] [CrossRef]
- Chen, D.; Sun, Y.; Zhao, L.; Qian, Y.; Zhu, R.; Liu, L.; Jia, L.; Dong, H.; Deng, J.; Deng, L. Development of Two Multiplex Real-Time PCR Assays for the Rapid Detection of RNA and DNA Viruses Associated with Gastroenteritis in Pediatric Patients. Pediat. Therapeut. 2014, 4, 208. [Google Scholar] [CrossRef]
- Logan, C.; O’Leary, J.J.; O’Sullivan, N. Real-time reverse transcription-PCR for detection of rotavirus and adenovirus as causative agents of acute viral gastroenteritis in children. J. Clin. Microbiol. 2006, 44, 3189–3195. [Google Scholar] [CrossRef]
- Philips, G.; Lopman, B.; Tam, C.C.; Iturriza-Gomara, M.; Brown, D.; Gray, J. Diagnosing rotavirus A associated IID: Using ELISA to identify a cut-off for real time RT-PCR. J. Clin. Vir. 2009, 44, 242–245. [Google Scholar] [CrossRef]
- Shen, X.X.; Qiu, F.Z.; Li, G.X.; Zhao, M.C.; Wang, J.; Chen, C.; Zhao, L.; Qi, J.J.; Liu, H.; Zhang, Y.; et al. A case control study on the prevalence of enterovirus in children samples and its association with diarrhea. Arch. Vir. 2018. [Google Scholar] [CrossRef]
- Tian, Y.; Chughtai, A.A.; Gao, Z.; Yan, H.; Chen, Y.; Liu, B.; Huo, D.; Jia, L.; Wang, Q.; MacIntyre, C.R. Prevalence and genotypes of group A rotavirus among outpatient children under five years old with diarrhea in Beijing, China, 2011–2016. BMC. Infect. Dis. 2018, 18, 497. [Google Scholar] [CrossRef]
- Chhabra, P.; Gregoricus, N.; Weinberg, G.A.; Halasa, N.; Chappell, J.; Hassan, F.; Selvarangan, R.; Mijatovic-Rustempasic, S.; Ward, M.L.; Bowen, M.; et al. Comparison of three multiplex gastrointestinal platforms for the detection of gastroenteritis viruses. J. Clin. Virol. 2017, 95, 66–71. [Google Scholar] [CrossRef]
- Can, V.D.; Kasuga, I.; Furumaj, H.; Katayama, H. Viability RT-qPCR Combined with Sodium Deoxycholate Pre-treatment for Selective Quantification of Infectious Viruses in Drinking Water Samples. Food. Environ. Virol. 2019, 11, 40–51. [Google Scholar] [CrossRef] [PubMed]

| Type | Pathogen | Number of Specimens | Source |
|---|---|---|---|
| Patients specimens | Adenovirus | 39 | Stool |
| Rotavirus | 349 | Stool | |
| Negative specimens | 297 | Stool | |
| Control specimens | Samonella Paratyphi A | 2 | Stool |
| Samonella Typhi | 1 | Stool | |
| Shigella Dysenteriae Type 2 | 5 | Stool | |
| Escherichia coli - 0:157:H7 | 1 | Culture | |
| Samonella enteritidis | 1 | Culture | |
| Shigella flexneri | 1 | Culture | |
| Shigella sonnei | 1 | Culture | |
| Vibrio cholera | 1 | Culture | |
| Camplyobacter jejuni | 1 | Culture | |
| Influenza B | 2 | NP swabs | |
| Influenza A/ H1N1 | 5 | NP swabs | |
| Influenza A/ H3N2 | 1 | NP swabs | |
| Picornavirus | 2 | NP swabs | |
| Aermonas hydrophila | 1 | Culture | |
| Neisseria meningitidis | 1 | Culture | |
| Streptococcus pyogenes | 2 | Culture | |
| Streptococcus pneumoniae | 2 | Culture | |
| Staphylococcus epidermidis | 1 | Culture | |
| Total | 716 |
| Target | Primers and Probe Sequences | Amp Size (BP) | NA Position | PCR Method | References |
|---|---|---|---|---|---|
| Adenovirus (hexon region) | FO: CAG GAC GCC TCG GRG TAY CTS AG RE: GGA GCC ACV GTG GGR TT PR: FAM-CCG GGT CTG GTG CAG TTT GCC CGC-BHQ | 103 | 17649–17752 * | rRT-PCR | Damen, M. et al. 2008 [17] |
| Rotavirus (NSP3 gene) | FO: CCA TCT WCA CRT RAC CCT CTA TGA G RE: GGT CAC ATA ACG CCC CTA TAG C PR: CY5-AGT TAA AAG CTA ACA CTG TCA AA-BHQ | 86 | 963–1049 | rRT-PCR | Zeng, S. et al. 2008 [18] |
| B. fragilis (16S rRNA gene) | FO: GAG AGG AAG GTC CC RE: CGC TAC TTG GCT GG PR: FAM-CCA TTG ACC AAT ATT CCT CAC TGC TGC CT-BHQ | 129 | 296–425 | RT-PCR | Layton, A. et al. 2006 [24] |
| MS2 (MS2-TM2JOE) | FO: GGC TGC TCG CGG ATA CC RE: TGA GGG AAT GTG GGA ACC G PR: JOE-ACC TCG GGT TTC CGT CTT GCT CGT-BHQ1 | 201 | 3166–3367 | rRT-PCR | Drier et al., 2005 [25] |
| A-Triplex Compared to Seegene | |||||||
| Seegene result for Adenovirus | Seegene result for Rotavirus | ||||||
| Triplex | Detected | Negative | Total | Triplex | Detected | Negative | Total |
| Adenovirus | 43 | 7 | 50 | Rotavirus | 106 | 4 | 110 |
| Negative | 0 | 319 | 319 | Negative | 1 | 258 | 259 |
| Total | 43 | 326 | 369 | Total | 107 | 262 | 369 |
| B-Triplex Compared to EM | |||||||
| EM result for Adenovirus | EM result for Rotavirus | ||||||
| Triplex | Detected | Negative | Total | Triplex | Detected | Negative | Total |
| Adenovirus | 37 | 23 | 60 | Rotavirus | 278 | 20 | 298 |
| Negative | 2 | 516 | 518 | Negative | 5 | 275 | 280 |
| Total | 39 | 539 | 578 | Total | 283 | 295 | 578 |
| C-Triplex Compared to ICT | |||||||
| ICT result for Rotavirus | |||||||
| Triplex | Detected | Negative | Total | ||||
| Rotavirus | 145 | 23 | 168 | ||||
| Negative | 15 | 137 | 152 | ||||
| Total | 160 | 160 | 320 | ||||
| A-Performance of Triplex Assay Against a Reference Standard Method | |||||
| Seegene | |||||
| Adenovirus | Rotavirus | ||||
| Triplex Accuracy | Formula | Estimate % (95% CI) | Estimate % (95% CI) | ||
| Sensitivity | TP/(TP + FN) × 100 | 100 (91.7–100) | 99.1 (94.9–99.8) | ||
| Specificity | TN/(FN + TN) × 100 | 97.8 (95.6–99.1) | 98.4 (96.1–99.6) | ||
| PPV | TP/(TP + FN) × 100 | 86 (74.7–92.7) | 96.3 (90.9–98.6) | ||
| NPV | TN/(FN + TN) × 100 | 100 | 99.6 (97.3–99.9) | ||
| B-Performance of Triplex Assay Against Two Non-Reference Standard Methods | |||||
| EM | ICT | ||||
| Adenovirus | Rotavirus | Rotavirus | |||
| Triplex Accuracy | Formula | Estimate % (95% CI) | Estimate % (95% CI) | Estimate % (95% CI) | |
| PPA | a/(a + c) × 100 | 94.8 (81.3–99.1) | 98.2 (95.6–99.3) | 90.6 (84.7–94.4) | |
| NPA | d/(b + d) × 100 | 95.7 (93.5–97.2) | 93.2 (89.5–95.7) | 85.6 (79.0–90.4) | |
| OPA | (a + d)/(a + b + c + d) × 100 | 95.6 (93.5–97.1) | 95.6 (93.5–97.1) | 88.1 (83.9–91.3) | |
| A-Adenovirus Results | ||||
| New Test | Non-Reference Standard Method | Total | Reference Standard Method | |
| Seegene | ||||
| Triplex | EM | + | − | |
| + | + | 32 | 32 | 0 |
| + | − | 15 | 8 | 7 |
| − | + | 1 | 0 | 1 |
| − | − | 286 | 0 | 286 |
| Total | 334 | 40 | 294 | |
| B-Rotavirus Results | ||||
| New Test | Non-reference Standard Method | Total | Reference Standard method | |
| Seegene | ||||
| Triplex | EM | + | − | |
| + | + | 97 | 97 | 0 |
| + | − | 7 | 3 | 4 |
| − | + | 2 | 0 | 2 |
| − | − | 228 | 0 | 228 |
| Total | 334 | 100 | 234 | |
| C-Rotavirus Results | ||||
| New Test | Non-Reference Standard Method | Total | Reference Standard Method | |
| Triplex | ICT | Seegene | ||
| + | − | |||
| + | + | 46 | 46 | 0 |
| + | − | 7 | 5 | 2 |
| − | + | 6 | 1 | 5 |
| − | − | 85 | 0 | 85 |
| Total | 144 | 52 | 92 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Higgins, R.R.; Peci, A.; Cardona, M.; Gubbay, J.B. Validation of a Laboratory-Developed Triplex Molecular Assay for Simultaneous Detection of Gastrointestinal Adenovirus and Rotavirus in Stool Specimens. Pathogens 2020, 9, 326. https://doi.org/10.3390/pathogens9050326
Higgins RR, Peci A, Cardona M, Gubbay JB. Validation of a Laboratory-Developed Triplex Molecular Assay for Simultaneous Detection of Gastrointestinal Adenovirus and Rotavirus in Stool Specimens. Pathogens. 2020; 9(5):326. https://doi.org/10.3390/pathogens9050326
Chicago/Turabian StyleHiggins, Rachel R., Adriana Peci, Mark Cardona, and Jonathan B. Gubbay. 2020. "Validation of a Laboratory-Developed Triplex Molecular Assay for Simultaneous Detection of Gastrointestinal Adenovirus and Rotavirus in Stool Specimens" Pathogens 9, no. 5: 326. https://doi.org/10.3390/pathogens9050326
APA StyleHiggins, R. R., Peci, A., Cardona, M., & Gubbay, J. B. (2020). Validation of a Laboratory-Developed Triplex Molecular Assay for Simultaneous Detection of Gastrointestinal Adenovirus and Rotavirus in Stool Specimens. Pathogens, 9(5), 326. https://doi.org/10.3390/pathogens9050326





