Coronavirus Disease Pandemic (COVID-19): Challenges and a Global Perspective
Abstract
:1. Introduction
2. Previous Human CoV Epidemics
2.1. SARS-CoV
2.2. MERS-CoV
2.3. SARS-CoV-2
3. SARS-CoV-2 Taxonomy, Structure, and Genomics
4. CoV Receptors
5. SARS-CoV-2 Persistence/Susceptibility on Different Surfaces
6. Phylogenomic Analysis of SARS-CoV-2 and Related CoVs
7. Epidemiology of SARS-CoV-2 Infection
7.1. Transmission
7.2. Incubation Period
7.3. Basic Reproduction Number (R0)
7.4. Population Susceptibility
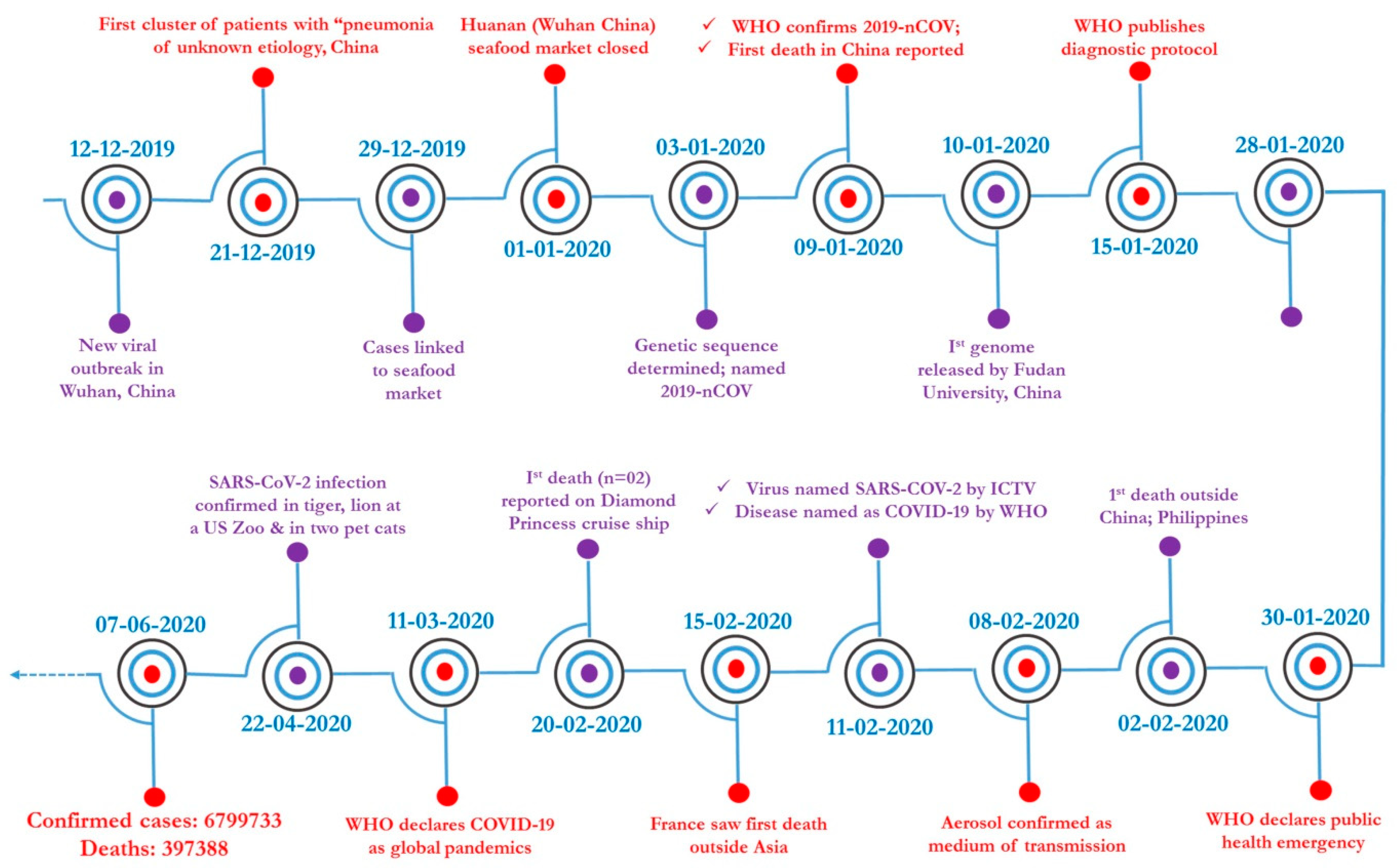
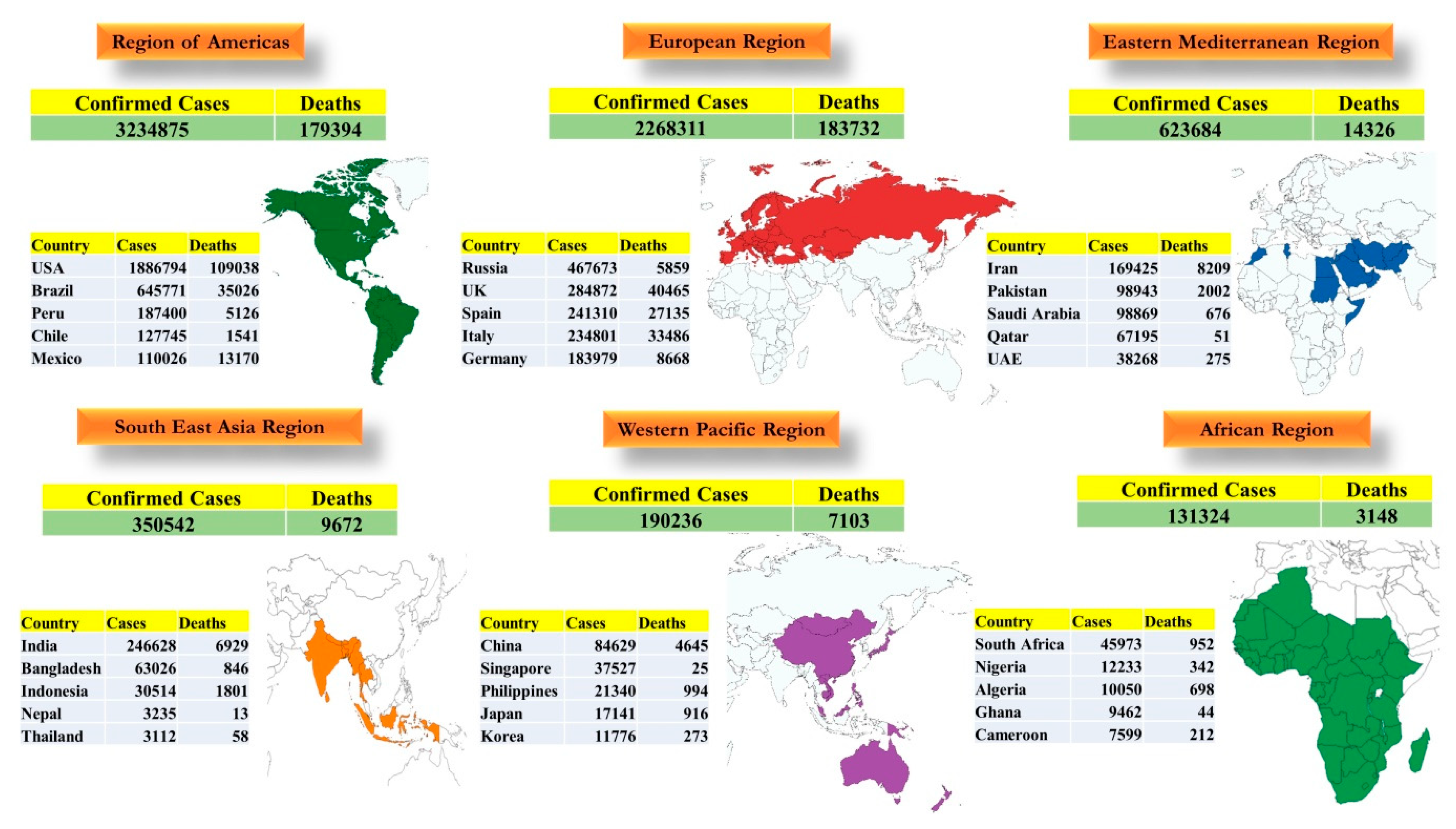
7.5. Global Scenario
7.6. Socio-Economic Impact
8. Clinical Profile
9. Clinical Pathology and Immunopathobiology
9.1. Clinical Pathology
9.2. Immunopathobiology
10. Animal Reservoirs
11. Animal Models in SARS-CoV-2 Research
12. Diagnosis of COVID-19
12.1. Sample Collection
12.2. Diagnostic Technologies
12.3. Artificial Intelligence in COVID-19 Diagnosis
13. Vaccines and Therapeutics
13.1. Vaccine Development
13.2. Development of Antiviral Therapeutics
13.3. Challenges of COVID-19 Vaccines and Therapeutics
14. Mitigation Strategies
15. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. Severe acute respiratory syndrome-related coronavirus: The species and its viruses—A statement of the Coronavirus Study Group. Nat. Microbiol. 2020, 55, 536–544. [Google Scholar] [CrossRef]
- Seven days in medicine: 8–14 January 2019. BMJ Case Rep. 2020. [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, P497–P506. [Google Scholar] [CrossRef] [Green Version]
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [PubMed]
- World Health Organization (WHO). Coronavirus Disease 2019 (COVID-19) Situation Report–93. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200422-sitrep-93-covid-19.pdf?sfvrsn=35cf80d7_4 (accessed on 23 April 2020).
- Wei, M.; Yuan, J.; Liu, Y.; Fu, T.; Yu, X.; Zhang, Z.J. Novel coronavirus infection in hospitalized infants under 1 year of age in China. J. Am. Med. Assoc. 2020, 323, 1313–1314. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Cai, H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir. Med. 2020, 8, e20. [Google Scholar] [CrossRef]
- Yu, W.-B.; Tang, G.-D.; Zhang, L.; Corlett, R. Decoding the evolution and transmissions of the novel pneumonia coronavirus (SARS-CoV-2) using the whole genomic data. Zool Res. 2020, 41, 247–257. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgson, J. The pandemic pipeline. Nat. Biotechnol. 2020, 38, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Rothe, C.; Schunk, M.; Sothmann, P.; Bretzel, G.; Froeschl, G.; Wallrauch, C.; Zimmer, T.; Thiel, V.; Janke, C.; Guggemos, W.; et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020, 382, 970–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.A.; Corman, V.M.; Jores, J.; Meyer, B.; Younan, M.; Liljander, A.; Bosch, B.J.; Lattwein, E.; Hilali, M.; Musa, B.E.; et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014, 20, 2093–2095. [Google Scholar] [CrossRef]
- Ellwanger, J.H.; Chies, J.A.B. Wind: A neglected factor in the spread of infectious diseases. Lancet Planet. Health 2018, 2, e475. [Google Scholar] [CrossRef] [Green Version]
- Lam, T.T.-Y.; Shum, M.H.-H.; Zhu, H.-C.; Tong, Y.-G.; Ni, X.-B.; Liao, Y.-S.; Wei, W.; Cheung, W.Y.-M.; Li, W.-J.; Li, L.-F.; et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature 2020. [Google Scholar] [CrossRef] [Green Version]
- Fung, T.S.; Liu, D.X. Human coronavirus: Host-pathogen interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Albarrak, A.M.; Omrani, A.S.; Albarrak, M.M.; Farah, M.E.; Almasri, M.; Muth, D.; Sieberg, A.; Meyer, B.; Assiri, A.M.; et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 2016, 62, 477–483. [Google Scholar] [CrossRef] [Green Version]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [Green Version]
- Vijayanand, P.; Wilkins, E.; Woodhead, M. Severe acute respiratory syndrome (SARS): A review. Clin. Med. (Lond.) 2004, 4, 152–160. [Google Scholar] [CrossRef]
- Ramadan, N.; Shaib, H. Middle East respiratory syndrome coronavirus (MERS-CoV): A review. Germs 2019, 9, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zheng, Y.; Gou, X.; Pu, K.; Chen, Z.; Guo, Q.; Ji, R.; Wang, H.; Wang, Y.; Zhou, Y. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: A systematic review and meta-analysis. Int. J. Infect. Dis. 2020, 94, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [Green Version]
- de Groot, R.J.; Baker, S.; Baric, R.; Enjuanes, L.; Gorbalenya, A.; Holmes, K.; Perlman, S.; Poon, L.; Rottier, P.; Talbot, P. Family coronaviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Academic Press: Cambridge, MA, USA, 2012; pp. 806–828. [Google Scholar]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Malik, Y.S.; Sircar, S.; Bhat, S.; Sharun, K.; Dhama, K.; Dadar, M.; Tiwari, R.; Chaicumpa, W. Emerging novel Coronavirus (2019-nCoV)-Current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020, 40, 68–76. [Google Scholar] [CrossRef]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Zhou, Y.; Liu, S.; Kou, Z.; Li, W.; Farzan, M.; Jiang, S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: Implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004, 324, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Le Coupanec, A.; Desforges, M.; Meessen-Pinard, M.; Dube, M.; Day, R.; Seidah, N.G.; Talbot, P.J. Cleavage of a neuroinvasive human respiratory virus spike glycoprotein by proprotein convertases modulates neurovirulence and virus spread within the central nervous system. PLoS Pathog. 2015, 11, e1005261. [Google Scholar] [CrossRef] [PubMed]
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nature 2016, 531, 118–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walls, A.C.; Tortorici, M.A.; Snijder, J.; Xiong, X.; Bosch, B.J.; Rey, F.A.; Veesler, D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. USA 2017, 114, 11157–11162. [Google Scholar] [CrossRef] [Green Version]
- Bassi, D.E.; Zhang, J.; Renner, C.; Klein-Szanto, A.J. Targeting proprotein convertases in furin-rich lung cancer cells results in decreased in vitro and in vivo growth. Mol. Carcinog. 2017, 56, 1182–1188. [Google Scholar] [CrossRef]
- Yang, Z.; Lasker, K.; Schneidman-Duhovny, D.; Webb, B.; Huang, C.C.; Pettersen, E.F.; Goddard, T.D.; Meng, E.C.; Sali, A.; Ferrin, T.E. UCSF Chimera, MODELLER, and IMP: An integrated modeling system. J. Struct. Biol. 2012, 179, 269–278. [Google Scholar] [CrossRef] [Green Version]
- Li, F. Receptor recognition and cross-species infections of SARS coronavirus. Antivir. Res. 2013, 100, 246–254. [Google Scholar] [CrossRef]
- Li, F. Receptor recognition mechanisms of coronaviruses: A decade of structural studies. J. Virol. 2015, 89, 1954–1964. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ace2 and tmprss2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K.; Li, W.; Moore, M.J.; Choe, H.; Farzan, M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004, 279, 3197–3201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, A.H.; Tomlinson, A.C.; Zhou, D.; Satkunarajah, M.; Chen, K.; Sharon, C.; Desforges, M.; Talbot, P.J.; Rini, J.M. Receptor-binding loops in alphacoronavirus adaptation and evolution. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Van Doremalen, N.; Bushmaker, T.; Munster, V. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Eurosurveillance 2013, 18, 20590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sizun, J.; Yu, M.; Talbot, P. Survival of human coronaviruses 229E and OC43 in suspension and after drying onsurfaces: A possible source ofhospital-acquired infections. J. Hosp. Infect. 2000, 46, 55–60. [Google Scholar] [CrossRef]
- Warnes, S.L.; Little, Z.R.; Keevil, C.W. Human coronavirus 229E remains infectious on common touch surface materials. mBio 2015, 6, e01697–e01715. [Google Scholar] [CrossRef] [Green Version]
- Rabenau, H.; Cinatl, J.; Morgenstern, B.; Bauer, G.; Preiser, W.; Doerr, H. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. 2005, 194, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Kratzel, A.; Todt, D.; V’kovski, P.; Steiner, S.; Gultom, M.L.; Thao, T.T.N.; Ebert, N.; Holwerda, M.; Steinmann, J.; Niemeyer, D.; et al. Efficient inactivation of SARS-CoV-2 by WHO-recommended hand rub formulations and alcohols. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kratzel, A.; Todt, D.; V’Kovski, P.; Steiner, S.; Gultom, M.; Thao, T.T.N.; Ebert, N.; Holwerda, M.; Steinmann, J.; Niemeyer, D.; et al. Inactivation of severe acute respiratory syndrome coronavirus 2 by WHO-Recommended hand rub formulations and alcohols. Emerg. Infect. Dis. 2020, 26, 1592–1595. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, N.; Havers, F.; Chen, T.; Song, Y.; Tu, W.; Li, L.; Cao, Y.; Liu, B.; Zhou, L.; Meng, L.; et al. Use of national pneumonia surveillance to describe influenza A(H7N9) virus epidemiology, China, 2004–2013. Emerg. Infect. Dis. 2013, 19, 1784–1790. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Guo, J.; Wang, C.; Luo, F.; Yu, X.; Zhang, W.; Li, J.; Zhao, D.; Xu, D.; Gong, Q.; et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. Lancet 2020, 395, 809–815. [Google Scholar] [CrossRef] [Green Version]
- Yeo, C.; Kaushal, S.; Yeo, D. Enteric involvement of coronaviruses: Is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020, 5, 335–337. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.-Y.; Chen, L.; Wang, M. Presumed asymptomatic carrier transmission of COVID-19. J. Am. Med. Assoc. 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [Green Version]
- Hindson, J. COVID-19: Faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 259. [Google Scholar] [CrossRef]
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020, 26, 502–505. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Du, R.H.; Li, B.; Zheng, X.S.; Yang, X.L.; Hu, B.; Wang, Y.Y.; Xiao, G.F.; Yan, B.; Shi, Z.L.; et al. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization (WHO). Coronavirus Disease 2019 (COVID-19). Situation Report–30. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200219-sitrep-30-covid-19.pdf?sfvrsn=3346b04f_2 (accessed on 8 April 2020).
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020, 27, taaa021. [Google Scholar] [CrossRef] [Green Version]
- Conti, P.; Younes, A. Coronavirus COV-19/SARS-CoV-2 Affects Women Less Than Men: Clinical Response to Viral Infection. J. Biol. Regul. Homeost. Agents. 2020, 34, 71. [Google Scholar] [CrossRef]
- Ronconi, G.; Teté, G.; Kritas, S.K.; Gallenga, C.E.; Caraffa, A.; Ross, R.; Conti, P. SARS-CoV-2, Which Induces COVID-19, Causes Kawasaki-Like Disease in Children: Role of Pro-Inflammatory and Anti-Inflammatory Cytokines. J. Biol. Regul. Homeost. Agents. 2020, 34. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Coronavirus Disease (COVID-19) Technical Guidance. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance (accessed on 28 March 2020).
- World Health Organization (WHO). Coronavirus Disease 2019 (COVID-19) Situation Report–2. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200122-sitrep-2-2019-ncov.pdf?sfvrsn=4d5bcbca_2 (accessed on 15 March 2020).
- World Health Organization (WHO). Coronavirus Disease 2019 (COVID-19) Situation Report–81. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200410-sitrep-81-covid-19.pdf?sfvrsn=ca96eb84_2 (accessed on 22 March 2020).
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Song, C.; Xu, C.; Jin, G.; Chen, Y.; Xu, X.; Ma, H.; Chen, W.; Lin, Y.; Zheng, Y. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci. China Life Sci. 2020, 63, 706–711. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention (CDC). Coronavirus Disease (COVID-19) Pregnancy and Breastfeeding. Available online: https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/pregnancy-breastfeeding.html (accessed on 7 June 2020).
- Singhal, T. A review of coronavirus disease-2019 (COVID-19). Indian J. Pediatr. 2020, 87, 281–286. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Deng, Y.; Li, W. Coronavirus disease 2019: What we know? J. Med. Virol. 2020, 92, 719–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Chan, K.H.; Peiris, J.S.; Lam, S.Y.; Poon, L.L.; Yuen, K.Y.; Seto, W.H. The Effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv. Virol. 2011, 2011, 734690. [Google Scholar] [CrossRef]
- Chan, J.F.; Yuan, S.; Kok, K.H.; To, K.K.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.; Poon, R.W.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.H.; Li, T.Y.; He, Z.C.; Ping, Y.F.; Liu, H.W.; Yu, S.C.; Mou, H.M.; Wang, L.H.; Zhang, H.R.; Fu, W.J.; et al. A pathological report of three COVID-19 cases by minimally invasive autopsies. J. Pathol. 2020, 49, 411–417. [Google Scholar]
- Xu, Y.H.; Dong, J.H.; An, W.M.; Lv, X.Y.; Yin, X.P.; Zhang, J.Z.; Dong, L.; Ma, X.; Zhang, H.J.; Gao, B.L. Clinical and computed tomographic imaging features of novel coronavirus pneumonia caused by SARS-CoV-2. J. Infect. 2020, 80, 394–400. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Liu, H.G.; Liu, W.; Liu, J.; Liu, K.; Shang, J.; Deng, Y.; Wei, S. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Int. J. Tuberc. Lung. Dis. 2020, 43, 203–208. [Google Scholar]
- Pan, Y.; Guan, H.; Zhou, S.; Wang, Y.; Li, Q.; Zhu, T.; Hu, Q.; Xia, L. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): A study of 63 patients in Wuhan, China. Eur. Radiol. 2020, 30, 3306–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, F.; Ye, T.; Sun, P.; Gui, S.; Liang, B.; Li, L.; Zheng, D.; Wang, J.; Hesketh, R.L.; Yang, L.; et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology 2020, 295, 715–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, J.; Li, J.; Li, X.; Qi, X. CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology 2020, 295, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Giwa, A.; Desai, A. Novel coronavirus COVID-19: An overview for emergency clinicians. Emerg. Med. Pract. 2020, 22, 1–28. [Google Scholar]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020, 53, 1689–1699. [Google Scholar] [CrossRef]
- Conti, P.; Ronconi, G.; Caraffa, A.; Gallenga, C.E.; Ross, R.; Frydas, I.; Kritas, S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVID-19 or SARS-CoV-2): Anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents 2020, 34, 1. [Google Scholar]
- Ehmann, K.Z.; Drosten, C.; Wendtner, C.; Zange, M.; Vollmar, P.; Rosina Ehmann, D.; Zwirglmaier, K.; Guggemos, M.; Seilmaier, M.; Niemeyer, D. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar]
- Peiris, J.; Lai, S.; Poon, L.; Guan, Y.; Yam, L.; Lim, W.; Nicholls, J.; Yee, W.; Yan, W.; Cheung, M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glass, W.G.; Subbarao, K.; Murphy, B.; Murphy, P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004, 173, 4030–4039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helms, J.; Tacquard, C.; Severac, F.; Leonard-Lorant, I.; Ohana, M.; Delabranche, X.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Gandet, F.F. High risk of thrombosis in patients with severe SARS-CoV-2 infection: A multicenter prospective cohort study. Intensive Care Med. 2020, 46, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Guan, W.; Chen, R.; Wang, W.; Li, J.; Xu, K.; Li, C.; Ai, Q.; Lu, W.; Liang, H. Cancer patients in SARS-CoV-2 infection: A nationwide analysis in china. Lancet Oncol. 2020, 21, 335–337. [Google Scholar] [CrossRef]
- Cameron, M.J.; Bermejo-Martin, J.F.; Danesh, A.; Muller, M.P.; Kelvin, D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res. 2008, 133, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef]
- Tetro, J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020, 22, 72–73. [Google Scholar] [CrossRef]
- Pedersen, S.F.; Ho, Y.-C. SARS-CoV-2: A storm is raging. J. Clin. Investig. 2020, 130. [Google Scholar] [CrossRef]
- Cao, X. COVID-19: Immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020, 22, 72–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zai, J.; Zhao, Q.; Nie, Q.; Li, Y.; Foley, B.T.; Chaillon, A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020, 92, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Wang, W.; Zhao, X.; Zai, J.; Li, X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020, 92, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Han, G.-Z. Pangolins harbor SARS-CoV-2-related coronaviruses. Trends Microbiol. 2020, 28, 515–517. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Veterinary Medical Association (AVMA). SARS-CoV-2 in Animals, Including Pets. Available online: https://www.avma.org/resources-tools/animal-health-and-welfare/covid-19/sars-cov-2-animals-including-pets (accessed on 10 April 2020).
- Callaway, E. Labs rush to study coronavirus in transgenic animals—Some are in short supply. Nature 2020, 579, 183. [Google Scholar] [CrossRef] [Green Version]
- Wentworth, D.E.; Gillim-Ross, L.; Espina, N.; Bernard, K.A. Mice susceptible to SARS coronavirus. Emerg. Infect. Dis. 2004, 10, 1293–1296. [Google Scholar] [CrossRef] [PubMed]
- Hogan, R.J.; Gao, G.; Rowe, T.; Bell, P.; Flieder, D.; Paragas, J.; Kobinger, G.P.; Wivel, N.A.; Crystal, R.G.; Boyer, J.; et al. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 2004, 78, 11416–11421. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.; Vogel, L.; Guarner, J.; Hayes, N.; Murphy, B.; Zaki, S.; Subbarao, K. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 2005, 79, 503–511. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.-I.; Kim, S.-G.; Kim, S.-M.; Kim, E.-H.; Park, S.-J.; Yu, K.-M.; Chang, J.-H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 2020, 27, C704–C709. [Google Scholar] [CrossRef]
- To, K.K.; Tsang, O.T.; Chik-Yan Yip, C.; Chan, K.H.; Wu, T.C.; Chan, J.M.C.; Leung, W.S.; Chik, T.S.; Choi, C.Y.; Kandamby, D.H.; et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foundation for Innovative New Diagnostic (FIND). COVID-19 Diagnostics Resource Centre. Available online: https://www.finddx.org/covid-19 (accessed on 12 April 2020).
- Sheridan, C. Fast, portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020, 38, 515–518. [Google Scholar] [CrossRef] [PubMed]
- CEepheid. Xpert®Xpress SARS-CoV-2. Available online: https://www.cepheid.com/coronavirus (accessed on 2 April 2020).
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IGM-IGG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J. SARS-CoV-2: An emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020, 16, 1678–1685. [Google Scholar] [CrossRef] [Green Version]
- Ai, J.W.; Zhang, Y.; Zhang, H.C.; Xu, T.; Zhang, W.H. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg. Microbes Infect. 2020, 9, 597–600. [Google Scholar] [CrossRef] [Green Version]
- Rao, A.S.S.; Vazquez, J.A. Identification of COVID-19 can be quicker through artificial intelligence framework using a mobile phone-based survey in the populations when cities/towns are under quarantine. Infect. Control Hosp. Epidemiol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Allam, Z.; Jones, D.S. On the coronavirus (COVID-19) outbreak and the smart city network: Universal data sharing standards coupled with artificial intelligence (AI) to benefit urban health monitoring and management. Healthcare 2020, 8, 46. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology 2020. [Google Scholar] [CrossRef]
- Santosh, K. AI-driven tools for coronavirus outbreak: Need of active learning and cross-population train/test models on multitudinal/multimodal data. J. Med. Syst. 2020, 44, 15. [Google Scholar] [CrossRef] [Green Version]
- Pang, J.; Wang, M.X.; Ang, I.Y.H.; Tan, S.H.X.; Lewis, R.F.; Chen, J.I.; Gutierrez, R.A.; Gwee, S.X.W.; Chua, P.E.Y.; Yang, Q.; et al. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): A systematic review. J. Clin. Med. 2020, 9, 623. [Google Scholar] [CrossRef] [Green Version]
- Le, T.T.; Andreadakis, Z.; Kumar, A.; Roman, R.G.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID -19 vaccine development landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306. [Google Scholar]
- ClinicalTrials.gov. Safety and Immunogenicity Study of 2019-nCoV Vaccine (mRNA-1273) for Prophylaxis SARS CoV-2 Infection (COVID-19). Available online: https://clinicaltrials.gov/ct2/show/NCT04283461 (accessed on 10 April 2020).
- Chinese Clinical Trial Registry. Available online: http://www.chictr.org.cn/showprojen.aspx?proj=51154,ChiCTR2000030906(accessed on 10 April 2020).
- Guo, D. Old weapon for new enemy: Drug repurposing for treatment of newly emerging viral diseases. Virol. Sin. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mifsud, E.J.; Hayden, F.G.; Hurt, A.C. Antivirals targeting the polymerase complex of influenza viruses. Antiviral Res. 2019, 169, 104545. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schafer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 222. [Google Scholar] [CrossRef] [Green Version]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A trial of lopinavir–ritonavir in adults hospitalized with severe COVID-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Gautret, P.; Lagier, J.C.; Parola, P.; Hoang, V.T.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 2020. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, J.; Bao, L.; Shi, Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020, 20, 398–400. [Google Scholar] [CrossRef]
- Rubbert-Roth, A.; Furst, D.E.; Nebesky, J.M.; Jin, A.; Berber, E. A review of recent advances using tocilizumab in the treatment of rheumatic diseases. Rheumatol. Ther. 2018, 5, 21–42. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, S.P.; Meng, S.; Wu, Y.J.; Mao, Y.P.; Ye, R.X.; Wang, Q.Z.; Sun, C.; Sylvia, S.; Rozelle, S.; Raat, H.; et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: A scoping review. Infect. Dis. Poverty 2020, 9, 29. [Google Scholar] [CrossRef] [Green Version]
- National Health Commission of People’s Republic of China (NHC). Notice on Printing and Distributing the Work Plan for Prevention and Control of Pneumonia Caused by Novel Coronavirus Infection in the Near Future. Available online: http://www.nhc.gov.cn/tigs/s7848/202001/808bbf75e5ce415aa19f74c78ddc653f.shtml (accessed on 11 April 2020).
- National Health Commission of People’s Republic of China (NHC). Notice on Prevention and Control of Novel Coronavirus Infection Pneumonia in the Elderly People. Available online: http://www.nhc.gov.cn/lljks/tggg/202001/96e82ba8a14d41b283da990d39771493.shtml (accessed on 31 January 2020).
- National Health Commission of People’s Republic of China (NHC). Notice on Further Prevention and Control of Pneumonia Caused by Novel Coronavirus Infection in Rural Areas. Available online: http://www.nhc.gov.cn/jkj/s3578/202001/f8d45f6af1d24ef18151c1d91cf8a028.shtml (accessed on 31 January 2020).
- World Health Organization (WHO). Advice on the Use of Masks in the Community, during Home Care and in Health Care Settings in the Context of the Novel Coronavirus 2019-nCoV Outbreak (Interim Guidance). Available online: WHO/2019-nCov/IPC_Masks/2020.3 (accessed on 8 April 2020).
- Radin, J.M.; Wineinger, N.E.; Topol, E.J.; Steinhubl, S.R.H. Fitbit-informed influenza forecasts. Lancet Digit. Health 2020. [Google Scholar] [CrossRef] [Green Version]
- Radin, J.M.; Wineinger, N.E.; Topol, E.J.; Steinhubl, S.R. Harnessing wearable device data to improve state-level real-time surveillance of influenza-like illness in the USA: A population-based study. Lancet Digit. Health 2020, 2, E85–E93. [Google Scholar] [CrossRef] [Green Version]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinman, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 114, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murdoch, D.R.; French, N.P. COVID-19: Another infectious disease emerging at the animal-human interface. N. Z. Med. J. 2020, 133, 12–15. [Google Scholar]
- Malik, Y.S.; Sircar, S.; Bhat, S.; Vinodhkumar, O.R.; Tiwari, R.; Sah, R.; Rabaan, A.A.; Rodriguez-Morales, A.J.; Dhama, K. Emerging coronavirus disease (COVID-19), a pandemic public health emergency with animal linkages: Current status update. Preprints 2020. [Google Scholar] [CrossRef]
- Dhama, K.; Patel, S.K.; Sharun, K.; Pathak, M.; Tiwari, R.; Yatoo, M.I.; Malik, Y.S.; Sah, R.; Rabaan, A.A.; Panwar, P.K.; et al. SARS-CoV-2: Jumping the species barrier, lessons from SARS and MERS, its zoonotic spillover, transmission to humans, preventive and control measures and recent developments to counter this pandemic virus. Preprints 2020. [Google Scholar] [CrossRef] [Green Version]
- Bonilla-Aldana, D.; Dhama, K.; Rodriguez-Morales, A. Editorial: Revisiting the one health approach in the context of COVID-19: A look into the ecology of this emerging disease. Adv. Animal Vet. Sci. 2020, 8, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Dhama, K.; Verma, A.; Tiwari, R.; Chakraborty, S.; Vora, K.; Sanjay, K.; Rajib, D.; Karthik, K.; Singh, R.; Munir, M.; et al. A perspective on applications of geographical information system (GIS); an advanced tracking tool for disease surveillance and monitoring in veterinary epidemiology. Adv. Animal Vet. Sci. 2013, 1, 14–24. [Google Scholar]
- World Health Organization (WHO). Battle against Respiratory Viruses (BRaVe) Initiative. Available online: https://www.who.int/influenza/patient_care/clinical/brave/en/ (accessed on 19 April 2020).
- Singh, R.K.; Dhama, K.; Chakraborty, S.; Tiwari, R.; Natesan, S.; Khandia, R.; Munjal, A.; Vora, K.S.; Latheef, S.K.; Karthik, K.; et al. Nipah virus: Epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies: A comprehensive review. Vet. Q. 2019, 39, 26–55. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Morales, A.; Bonilla-Aldana, D.; Tiwari, R.; Sah, R.; Rabaan, A.; Dhama, K. COVID-19 an emerging coronavirus infection: Current scenario and recent developments—An Overview. J. Pure Appl. Microbiol. 2020, 14, 6150. [Google Scholar] [CrossRef] [Green Version]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-CoV: A comparative overview. Infez. Med. 2020, 28, 174–184. [Google Scholar] [PubMed]
- Rabaan, A.A.; Al-Ahmed, S.H.; Sah, R.; Tiwari, R.; Yatoo, M.I.; Patel, S.K.; Pathak, M.; Malik, Y.S.; Dhama, K.; Singh, K.P.; et al. SARS-CoV-2/COVID-19 and advances in developing potential therapeutics and vaccines to counter this emerging pandemic virus: A review. Preprints 2020. [Google Scholar] [CrossRef]
- Munjal, A.; Khandia, R.; Dhama, K.; Sachan, S.; Karthik, K.; Tiwari, R.; Malik, Y.S.; Kumar, D.; Singh, R.K.; Iqbal, H.M.N.; et al. Advances in developing therapies to combat Zika Virus: Current knowledge and future perspectives. Front. Microbiol. 2017, 8, 1469. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Sharun, K.; Tiwari, R.; Dadar, M.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W. COVID-19, an emerging coronavirus infection: Advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vaccin. Immunother. 2020. [Google Scholar] [CrossRef] [Green Version]
- Dhama, K.; Karthik, K.; Khandia, R.; Chakraborty, S.; Munjal, A.; Latheef, S.K.; Kumar, D.; Ramakrishnan, M.A.; Malik, Y.S.; Singh, R.; et al. Advances in designing and developing vaccines, drugs, and therapies to counter Ebola virus. Front. Immunol. 2018, 9, 1803. [Google Scholar] [CrossRef]
- Chatterjee, P.; Nagi, N.; Agarwal, A.; Das, B.; Banerjee, S.; Sarkar, S.; Gupta, N.; Gangakhedkar, R.R. The 2019 novel coronavirus disease (COVID-19) pandemic: A review of the current evidence. Indian J. Med. Res. 2020, 151, 147. [Google Scholar] [CrossRef]
- Conti, P.; Gallenga, C.E.; Tetè, G.; Caraffa, A.; Ronconi, G.; Younes, A.; Toniato, E.; Ross, R.; Kritas, S.K. How to reduce the likelihood of coronavirus-19 (CoV-19 or SARS-CoV-2) infection and lung inflammation mediated by IL-1. J. Biol. Regul. Homeost. Agents. 2020, 34. [Google Scholar] [CrossRef]
- El Zowalaty, M.E.; Järhult, J.D. From SARS to COVID-19: A previously unknown SARS- related coronavirus (SARS-CoV-2) of pandemic potential infecting humans—Call for a one health approach. One Health 2020, 9, 100124. [Google Scholar] [CrossRef]



| Features | SARS | MERS | COVID-19 |
|---|---|---|---|
| Causative agent | SARS-CoV | MERS-CoV | SARS-CoV-2 |
| Incubation period | 2–10 days | 2–10 days | 2–14 days |
| The median age of infected human cases | 65 years | 50 years | 59 years |
| Source of origin | Bats, civet cats | Bats, camel | Seafood, bats, pangolin (proposed) |
| Transmission | Animal–human Human–human Zoonotic disease | Animal–human Human–human Zoonotic disease | Animal–human Human–human Human–animal Zoonotic disease |
| Speed of spread | Moderate | Low | High |
| Seasonal occurrence | Winter (Dec–Jan) | Summer (May–July) | Winter (Dec–Jan) |
| Place of Origin | Guangdong, China | Jeddah, Saudi Arabia | Wuhan, China |
| First incidence | 16 November 2002 | 13 June 2012 | 7 December 2019 |
| The last case reported and present status | 5 July 2003 | 18 February 2020 (Ongoing) | Ongoing |
| Total number of cases | 8422 | 2496 | 1,434,426 as of 8 April 2020 |
| Overall fatality | 916 (10.87%) | 868 (34.77%) (as of now) | 82,220 (5.73%) as of 8 April 2020 |
| No. Countries affected | 31 | 27 | 212 (till now) |
| Intermediate host | Paguma larvata | Camelus dromedaries | Pangolin, Mink (possible) |
| Definitive host | Rhinolophus sinicus | Pipistrellus hesperidus | Rhinolophus affinis (possible) |
| Taxonomy | Betacoronavirus (Serbecovirus) | Betacoronavirus (Merbecovirus) | Betacoronavirus (Serbecovirus) |
| Genome length (bases) | 29751 | 30119 | 29903 |
| Major Regional distribution | Guangdong province of southern China, and later to western pacific countries | Saudi Arabia, followed by UAE and Republic of Korea | Hubei, especially, Wuhan in China, followed by worldwide |
| Treatment/Vaccine | Glucocorticoid and Interferon | No effective approved treatment or vaccine | Lopinavir/Ritonavir (in testing) |
| Receptor | Angiotensin-converting enzyme-2 (ACE-2) | Dipeptidyl peptidase 4 (DPP4) | ACE-2 |
| Genus | CoVs | Receptors |
|---|---|---|
| Alphacoronavirus | HCoV-NL63 | ACE-2 |
| TGEV | APN | |
| PRCV | APN | |
| Betacoronavirus | MHV | CEACAM1 |
| BCoV | N-acetyl-9-O-acetylneuraminic acid | |
| MERS | DPP4 (CD26) | |
| SARS | ACE-2 | |
| Gammacoronavirus | IBV | α2,3-linked sialic acid (attachment factor) |
| Deltacoronavirus | PDCoV | APN |
| CoVs | Strain Name/ Accession Number | % Nucleotide Sequence Identity | % Protein Sequence Identity |
|---|---|---|---|
| (a)Whole Genome-Based Comparison (29.9kb) | |||
| South Korea | SNU01/MT039890 | 99 | 99.9 |
| Australia | VIC01/MT007544 | 99 | 99.9 |
| Nepal | 61-TW/MT072688 | 99–100 | 100 |
| Taiwan | NTU02/MT066176 | 99–100 | 100 |
| USA | USA-CA9/MT118835 | 99–100 | 100 |
| Japan | WK-501/LC522974 | 99–100 | 100 |
| (b) Spike Protein-Based Comparison (~3800 bases) | |||
| Pangolin CoV | MP789/MT084071 | 89.7 | 89.78 |
| Bat_SL_CoVs | bat-SL-CoVZC45/MG772933 bat-SL-CoVZXC21/MG772934 | 77.5–78.3 | 81.2–81.8 |
| Bat_CoV | RaTG13/MN996532 | 93.1 | 97.69 |
| Bat_SARS_CoVs | WIV16/KT444582 BtRs-BetaCoV/YN2018C/MK211377 | 69.8–73.9 | 72.3–78.1 |
| SARS_CoVs | NS-1/AY508724 GD01/AY278489 | 73.9 | 77 |
| MERS_CoVs | HCoV-EMC/JX869059 England 1/NC_038294 | 21.8–21.9 | 26.8 |
| CRCoVs | CRCVK39/EU983107 Respiratory/AY150272 | 23.1–23.2 | 28.8–29.1 |
| BCoVs | CAMAGUEY/HE616741 HT317/MK046011 | 18.2–22.7 | 16.9–28.7 |
| EqCoV | Obihiro12-1/LC061273 | 18.3 | 29.2 |
| HCoV | OC43/S62886 | 18.1–18.7 | 26.8–28.0 |
| Bat_CoV | Zhejiang2013/KF636752 | 33.6 | 39.0 |
| Bat_CoVs | KHU9-1/EF065513 GCCDC1 356/KU762338 | 24.3–25.1 | 29.1 |
| Company | Assay | Targets | Regulatory Status |
|---|---|---|---|
| Centers for Disease Control and Prevention (CDC) | CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel (CDC) | N1, N2 and RNase P (control) | USA FDA-EUA |
| Roche Molecular Systems, Inc. (RMS) | cobas SARS-CoV-2 | ORF1ab, E gene, RNase P (control) | USA FDA-EUA |
| Thermo Fisher Scientific, Inc. | TaqPath COVID-19 Combo Kit | ORF1ab, N gene, S gene, MS2 (control) | USA FDA-EUA |
| Primerdesign Ltd. | Primerdesign Ltd. COVID-19 genesig Real-Time PCR assay | RdRp gene | USA FDA-EUA |
| Abbott Molecular | Abbott RealTime SARS-CoV-2 assay | RdRp and N genes | USA FDA-EUA |
| Cepheid | Xpert Xpress SARS-CoV-2 test | N2 and E genes | USA FDA-EUA |
| DAAN Gene Co., Ltd. of Sun Yat-sen University | Detection Kit for 2019 Novel Coronavirus (2019-nCoV) RNA (PCR-Fluorescence Probing) | ORF1ab and N gene | China EUA |
| Seegene, Inc. | Allplex 2019-nCoV assay | E, RdRp and N genes | Korea EUA |
| Vaccine/Drug Candidates | Developer | Status | Remarks |
|---|---|---|---|
| Vaccines candidates | |||
| mRNA-1273 | Moderna/NIAID | Phase 1 (NCT04283461) | Clinical trials started at Kaiser Permanente Washington Health Research Institute in Seattle, USA |
| Ad5-nCov | CanSino Biological Inc. and Beijing Institute of Biotechnology | Phase 1 (ChiCTR2000030906) | Clinical trials recently started at Tongji Hospital in Wuhan, China |
| INO-4800 | Inovio Pharmaceuticals | Phase I (NCT04336410) | DNA plasmid encoding S protein delivered by electroporation |
| LV-SMENP-DC | Shenzhen Geno-Immune Medical Institute | Phase I (NCT04276896) | Dendritic cells modified with lentiviral vector and expressing synthetic minigene based on domains of selected viral proteins |
| Pathogen-specific aAPC | Shenzhen Geno-Immune Medical Institute | Phase I (NCT04299724) | Artificial antigen-presenting cells (aAPCs) modified with lentiviral vector and expressing synthetic minigene based on domains of selected viral proteins |
| Drug candidates | |||
| Favilavir | Hisun Pharmaceutical, China | Approved by NMPA, China | Inhibition of RNA-dependent RNA polymerase of SARS-CoV-2 |
| Hydroxychloroquine and azithromycin | USA | Clinical trials are under progress | Both have shown in vitro activity against SARS-CoV-2 |
| Remdesivir | Gilead Sciences, USA | Clinical trials are under progress | Developed for treatment of Ebola virus infection |
| Lopinavir + Ritonavir | AbbVie, USA | Further research is under process | Developed to treat HIV-1 infections |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malik, Y.S.; Kumar, N.; Sircar, S.; Kaushik, R.; Bhat, S.; Dhama, K.; Gupta, P.; Goyal, K.; Singh, M.P.; Ghoshal, U.; et al. Coronavirus Disease Pandemic (COVID-19): Challenges and a Global Perspective. Pathogens 2020, 9, 519. https://doi.org/10.3390/pathogens9070519
Malik YS, Kumar N, Sircar S, Kaushik R, Bhat S, Dhama K, Gupta P, Goyal K, Singh MP, Ghoshal U, et al. Coronavirus Disease Pandemic (COVID-19): Challenges and a Global Perspective. Pathogens. 2020; 9(7):519. https://doi.org/10.3390/pathogens9070519
Chicago/Turabian StyleMalik, Yashpal Singh, Naveen Kumar, Shubhankar Sircar, Rahul Kaushik, Sudipta Bhat, Kuldeep Dhama, Parakriti Gupta, Kapil Goyal, Mini P Singh, Ujjala Ghoshal, and et al. 2020. "Coronavirus Disease Pandemic (COVID-19): Challenges and a Global Perspective" Pathogens 9, no. 7: 519. https://doi.org/10.3390/pathogens9070519
APA StyleMalik, Y. S., Kumar, N., Sircar, S., Kaushik, R., Bhat, S., Dhama, K., Gupta, P., Goyal, K., Singh, M. P., Ghoshal, U., El Zowalaty, M. E., O. R, V., Yatoo, M. I., Tiwari, R., Pathak, M., Patel, S. K., Sah, R., Rodriguez-Morales, A. J., Ganesh, B., ... Singh, R. K. (2020). Coronavirus Disease Pandemic (COVID-19): Challenges and a Global Perspective. Pathogens, 9(7), 519. https://doi.org/10.3390/pathogens9070519












