Identification of Potential Drug Targets in Helicobacter pylori Using In Silico Subtractive Proteomics Approaches and Their Possible Inhibition through Drug Repurposing
Abstract
:1. Introduction
2. Results
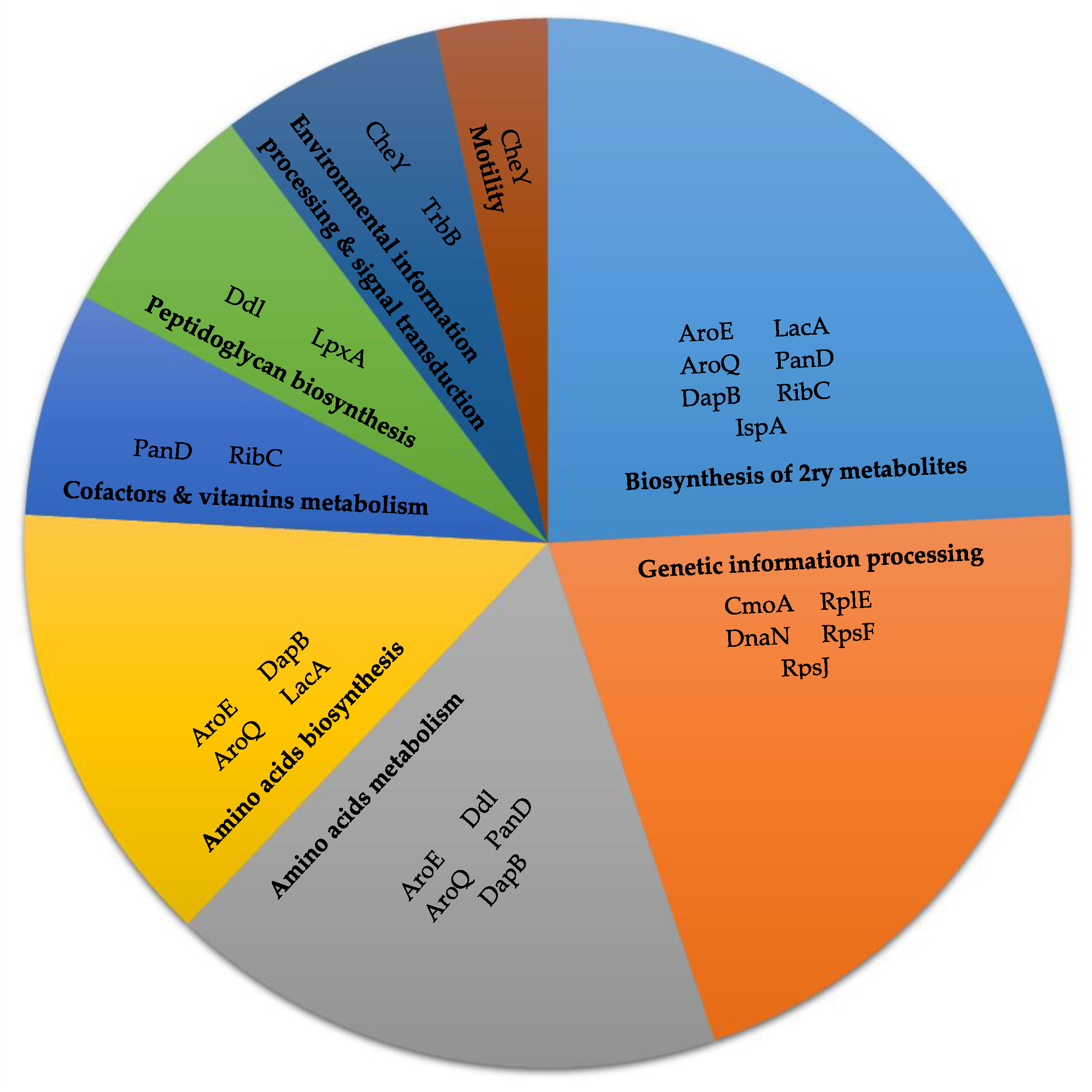
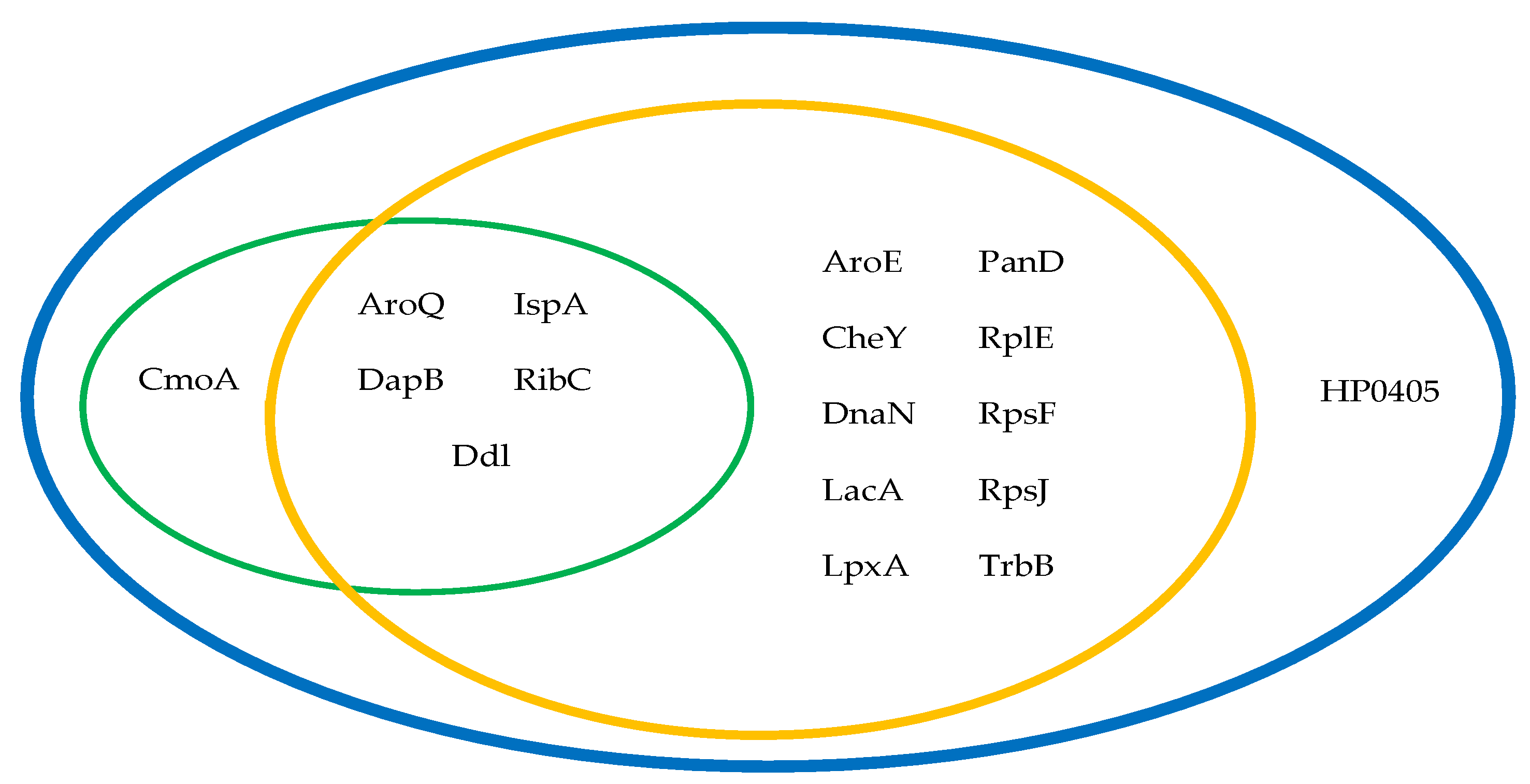
2.1. Chokepoint Proteins Analysis
2.2. Metabolic Pathway Analysis
2.3. Essential Proteins Analysis
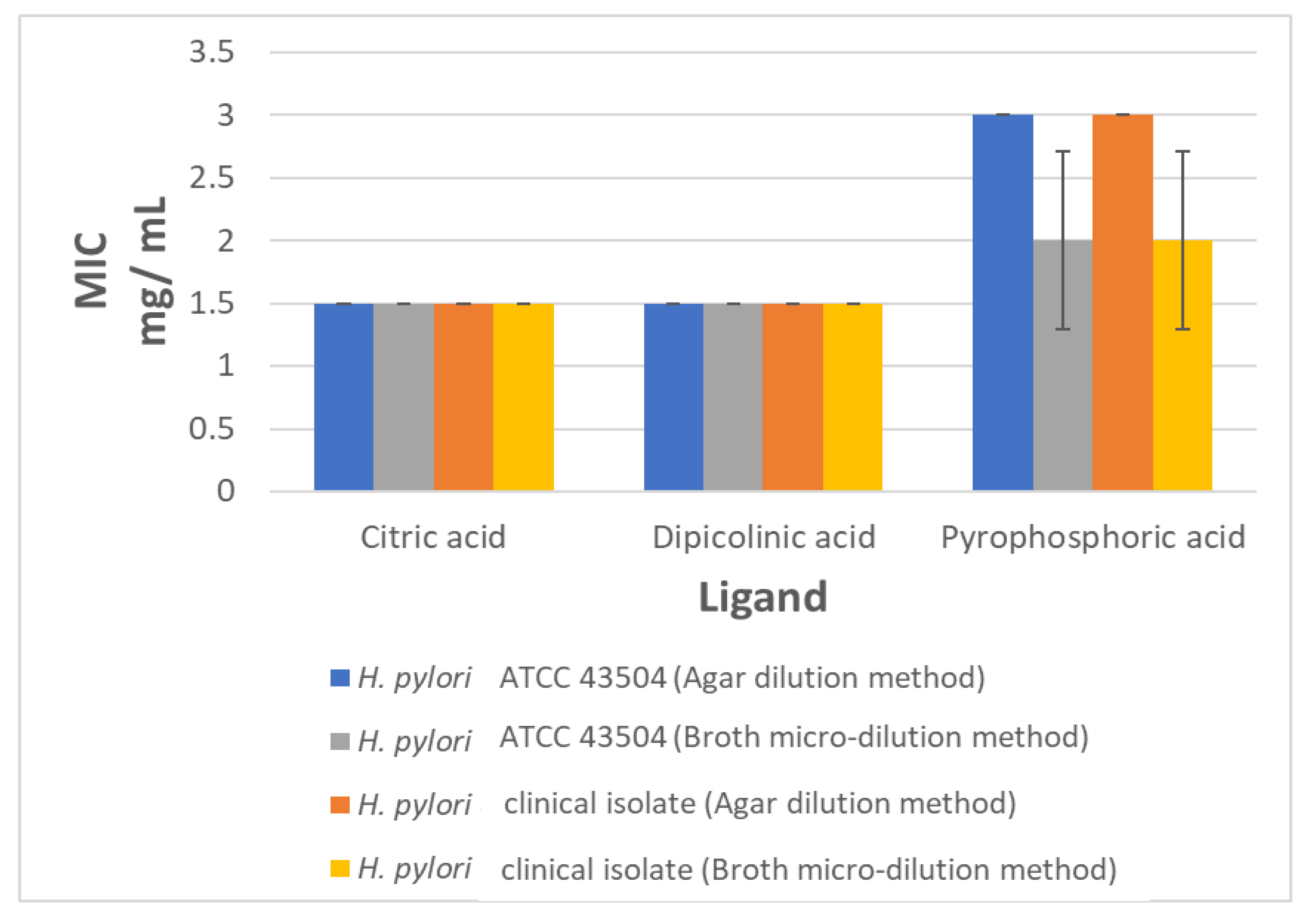
2.4. The In Vitro Anti-Helicobacter Activity of Selected Ligands
3. Discussion
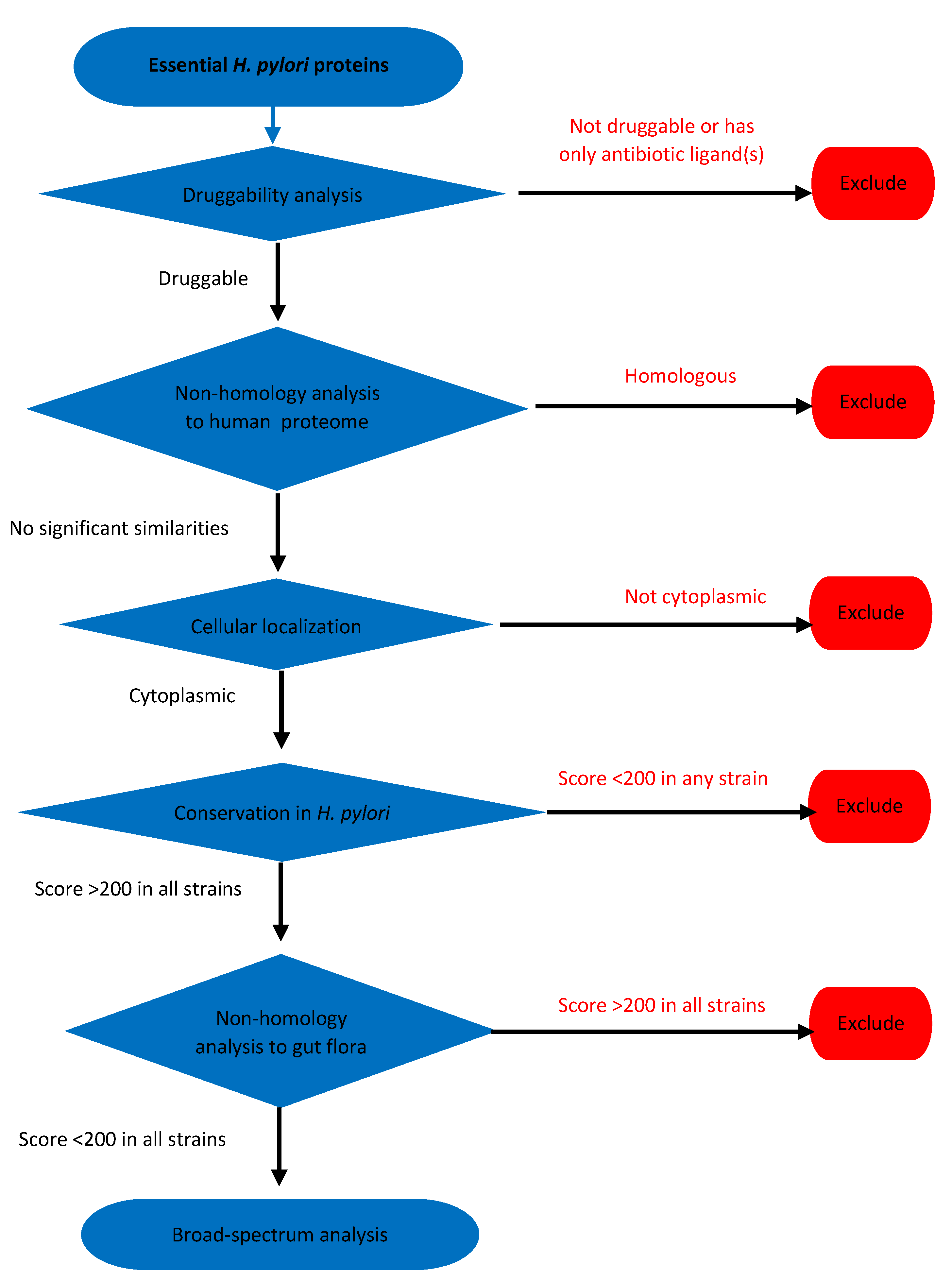
4. Materials and Methods
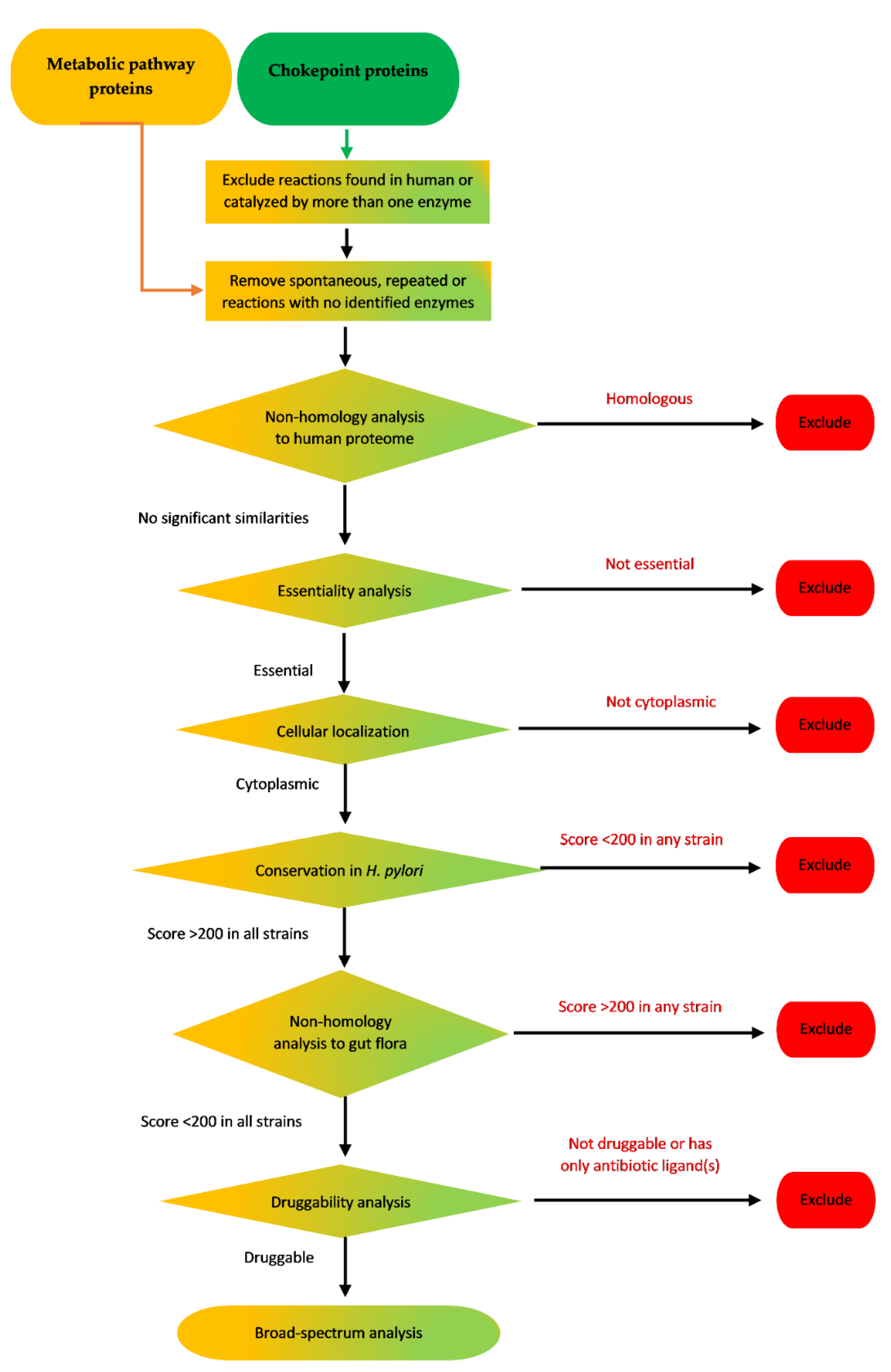
4.1. Analysis of Chokepoint Proteins and Metabolic Pathway Proteins
4.1.1. Retrieval of Chokepoint Proteins of H. pylori
4.1.2. Retrieval of Proteins Involved in H. pylori Metabolic Pathways
4.1.3. Non-Homology Analysis to Human Proteins
4.1.4. Essentiality Analysis
4.1.5. Prediction of Sub-Cellular Localization
4.1.6. Conservation in H. pylori
4.1.7. Non-Homology to Proteome of Common Gut Flora Organisms
4.1.8. Druggability Analysis
4.1.9. Checking for Broad-Spectrum Targeting
4.2. Essential Proteins Analysis
4.3. Determination of MIC
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Duck, W.M.; Sobel, J.; Pruckler, J.M.; Song, Q.; Swerdlow, D.; Friedman, C.; Sulka, A.; Swaminathan, B.; Taylor, T.; Hoekstra, M.; et al. Antimicrobial Resistance Incidence and Risk Factors among Helicobacter pylori–Infected Persons, United States. Emerg. Infect. Dis. 2004, 10, 1088–1094. [Google Scholar] [CrossRef]
- Guevara, B.; Cogdill, A.G. Helicobacter pylori: A Review of Current Diagnostic and Management Strategies. Dig. Dis. Sci. 2020, 65, 1917–1931. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Schistosomes, Liver Flukes and Helicobacter Pylori; International Agency for Research on Cancer: Lyon, France, 1994; Volume 61. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, A.; Dubey, J.; Hegde, R.R.; Rastogi, V.; Pandit, J.K. Helicobacter pylori: Past, current and future treatment strategies with gastroretentive drug delivery systems. J. Drug Target. 2016, 24, 897–915. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, A.; Liou, J.; Gisbert, J.P.; O’Morain, C. Review: Treatment of Helicobacter pylori Infection. Helicobacter 2019, 24, e12640. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.K.; Ahn, J.Y.; Won, S.H.; Jung, K.; Na, H.K.; Jung, K.W.; Kim, H.; Lee, J.H.; Choi, K.D.; Song, H.J.; et al. Eradication rate of Helicobacter pylori reinfection in Korea: A retrospective study. J. Gastroenterol. Hepatol. 2019, 34, 1696–1702. [Google Scholar] [CrossRef]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. Available online: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 1 May 2020).
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlen, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2019, 18, 275–285. [Google Scholar] [CrossRef] [Green Version]
- Miró-Canturri, A.; Ayerbe-Algaba, R.; Smani, Y. Drug Repurposing for the Treatment of Bacterial and Fungal Infections. Front. Microbiol. 2019, 10, 41. [Google Scholar] [CrossRef]
- Cai, J.; Han, C.; Hu, T.; Zhang, J.; Wu, D.; Wang, F.; Liu, Y.; Ding, J.; Chen, K.; Yue, J.; et al. Peptide deformylase is a potential target for Anti-Helicobacter pyloridrugs: Reverse docking, enzymatic assay, and X-ray crystallography validation. Protein Sci. 2006, 15, 2071–2081. [Google Scholar] [CrossRef] [Green Version]
- Rout, S.; Patra, N.P.; Mahapatra, R.K. An in silico strategy for identification of novel drug targets against Plasmodium falciparum. Parasitol. Res. 2017, 116, 2539–2559. [Google Scholar] [CrossRef] [PubMed]
- Fields, F.R.; Lee, S.W.; McConnell, M.J. Using bacterial genomes and essential genes for the development of new antibiotics. Biochem. Pharmacol. 2017, 134, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Wadood, A.; Ahmed, N.; Shah, L.; Ahmad, A.; Hassan, H.; Shams, S. In-silico drug design: An approach which revolutionarised the drug discovery process. OA Drug Des. Deliv. 2013, 1, 3. [Google Scholar] [CrossRef]
- Chong, C.E.; Lim, B.-S.; Nathan, S.; Mohamed, R. In silico analysis of Burkholderia pseudomallei genome sequence for potential drug targets. Silico Boil. 2006, 6, 341–346. [Google Scholar]
- Sarangi, A.N.; Aggarwal, R.; Rahman, Q.; Trivedi, N. Subtractive Genomics Approach for in Silico Identification and Characterization of Novel Drug Targets in Neisseria Meningitides Serogroup B. J. Comput. Sci. Syst. Boil. 2009, 2, 255–258. [Google Scholar] [CrossRef]
- Shahid, F.; Ashfaq, U.A.; Saeed, S.; Munir, S.; Almatroudi, A.; Khurshid, M. In Silico Subtractive Proteomics Approach for Identification of Potential Drug Targets in Staphylococcus saprophyticus. Int. J. Environ. Res. Public Health 2020, 17, 3644. [Google Scholar] [CrossRef]
- Solanki, V.; Tiwari, V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci. Rep. 2018, 8, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Uddin, R.; Jamil, F. Prioritization of potential drug targets against P. aeruginosa by core proteomic analysis using computational subtractive genomics and Protein-Protein interaction network. Comput. Boil. Chem. 2018, 74, 115–122. [Google Scholar] [CrossRef]
- Uddin, R.; Siraj, B.; Rashid, M.; Khan, A.; Halim, S.A.; Al-Harrasi, A. Genome Subtraction and Comparison for the Identification of Novel Drug Targets against Mycobacterium avium subsp. hominissuis. Pathogens 2020, 9, 368. [Google Scholar] [CrossRef]
- Wadood, A.; Jamal, A.; Riaz, M.; Khan, A.; Uddin, R.; Jelani, M.; Azam, S.S. Subtractive genome analysis for in silico identification and characterization of novel drug targets in Streptococcus pneumonia strain JJA. Microb. Pathog. 2018, 115, 194–198. [Google Scholar] [CrossRef]
- Dutta, A.; Singh, S.K.; Ghosh, P.; Mukherjee, R.; Mitter, S.; Bandyopadhyay, D. In silico identification of potential therapeutic targets in the human pathogen Helicobacter pylori. Silico Boil. 2006, 6, 43–47. [Google Scholar]
- Neelapu, N.R.R.; Pavani, T. Identification of novel drug targets in HpB38, HpP12, HpG27, Hpshi470, HpSJM180 strains of Helicobacter pylori: An in silico approach for therapeutic intervention. Curr. Drug Targets 2013, 14, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Neelapu, N.R.R.; Mutha, N.; Akula, S. Identification of Potential Drug Targets in Helicobacter pylori Strain HPAG1 by in silico Genome Analysis. Infect. Disord. Drug Targets 2015, 15, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.; Maganti, L.; Ghoshal, N.; Dutta, C. In silico quest for putative drug targets in Helicobacter pylori HPAG1: Molecular modeling of candidate enzymes from lipopolysaccharide biosynthesis pathway. J. Mol. Model. 2012, 18, 1855–1866. [Google Scholar] [CrossRef]
- Pasala, C.; Chilamakuri, C.S.R.; Katari, S.K.; Nalamolu, R.M.; Bitla, A.R.; Umamaheswari, A. An in silico study: Novel targets for potential drug and vaccine design against drug resistant H. pylori. Microb. Pathog. 2018, 122, 156–161. [Google Scholar] [CrossRef]
- Yeh, I.; Hanekamp, T.; Tsoka, S.; Karp, P.D.; Altman, R.B. Computational Analysis of Plasmodium falciparum Metabolism: Organizing Genomic Information to Facilitate Drug Discovery. Genome Res. 2004, 14, 917–924. [Google Scholar] [CrossRef] [Green Version]
- Raman, K.; Yeturu, K.; Chandra, N.R. targetTB: A target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. BMC Syst. Boil. 2008, 2, 109. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearson, W.R. Selecting the Right Similarity-Scoring Matrix. Curr. Protoc. Bioinform. 2013, 43, 3.5.1–3.5.9. [Google Scholar] [CrossRef] [PubMed]
- Butt, A.M.; Tahir, S.; Nasrullah, I.; Idress, M.; Lu, J.; Tong, Y. Mycoplasma genitalium: A comparative genomics study of metabolic pathways for the identification of drug and vaccine targets. Infect. Genet. Evol. 2012, 12, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Damte, D.; Suh, J.-W.; Lee, S.-J.; Yohannes, S.B.; Hossain, A.; Park, S.-C. Putative drug and vaccine target protein identification using comparative genomic analysis of KEGG annotated metabolic pathways of Mycoplasma hyopneumoniae. Genomics 2013, 102, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Jadhav, A.; Shanmugham, B.; Rajendiran, A.; Pan, A. Unraveling novel broad-spectrum antibacterial targets in food and waterborne pathogens using comparative genomics and protein interaction network analysis. Infect. Genet. Evol. 2014, 27, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lin, Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 2009, 37, D455–D458. [Google Scholar] [CrossRef] [Green Version]
- Ramos, P.I.P.; Porto, D.A.F.D.; Lanzarotti, E.; Sosa, E.J.; Burguener, G.; Pardo, A.; Klein, C.C.; Sagot, M.-F.; Vasconcelos, A.T.R.; Gales, A.C.; et al. An integrative, multi-omics approach towards the prioritization of Klebsiella pneumoniae drug targets. Sci. Rep. 2018, 8, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Barh, D.; Tiwari, S.; Jain, N.; Ali, A.; Santos, A.R.; Misra, A.N.; Azevedo, V.; Kumar, A. In silico subtractive genomics for target identification in human bacterial pathogens. Drug Dev. Res. 2010, 72, 162–177. [Google Scholar] [CrossRef]
- Benites, J.; Toledo, H.; Salas, F.; Guerrero, A.; Rios, D.; Valderrama, J.A.; Calderon, P.B. In Vitro Inhibition ofHelicobacter pyloriGrowth by Redox Cycling Phenylaminojuglones. Oxidative Med. Cell. Longev. 2018, 2018, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Satuluri, S.H.; Katari, S.K.; Pasala, C.; Amineni, U. Novel and potent inhibitors for dihydropteroate synthase of Helicobacter pylori. J. Recept. Signal Transduct. 2020, 40, 246–256. [Google Scholar] [CrossRef]
- Song, Z.; Fu, W.; Zhou, L. Cefuroxime, levofloxacin, esomeprazole, and bismuth as first-line therapy for eradicating Helicobacter pylori in patients allergic to penicillin. BMC Gastroenterol. 2019, 19, 132. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Belenky, P.; Yang, J.H.; Macdonald, I.C.; Martell, J.D.; Takahashi, N.; Chan, C.T.Y.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100–E2109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Xiao, H.; Bonanno, J.B.; Kalyanaraman, C.; Brown, S.; Tang, X.; Al-Obaidi, N.; Patskovsky, Y.; Babbitt, P.C.; Jacobson, M.P.; et al. Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature 2013, 498, 123–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, S.; Reader, J.S. tRNAs as Antibiotic Targets. Int. J. Mol. Sci. 2015, 16, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Park, S.Y.; Jeong, S.J.; Jung, D.H.; Kim, J.-H.; Jeong, S.H.; Kang, I.-M.; Song, Y.G. Can Aminoglycosides Be Used as a New Treatment for Helicobacter pylori? In vitro Activity of Recently Isolated Helicobacter pylori. Infect. Chemother. 2019, 51, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Foynes, S.; Dorrell, N.; Ward, S.J.; Stabler, R.A.; McColm, A.A.; Rycroft, A.N.; Wren, B.W. Helicobacter pylori Possesses Two CheY Response Regulators and a Histidine Kinase Sensor, CheA, Which Are Essential for Chemotaxis and Colonization of the Gastric Mucosa. Infect. Immun. 2000, 68, 2016–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walia, G.; Kumar, P.; Surolia, A. The Role of UPF0157 in the Folding of M. tuberculosis Dephosphocoenzyme A Kinase and the Regulation of the Latter by CTP. PLoS ONE 2009, 4, e7645. [Google Scholar] [CrossRef] [Green Version]
- Spry, C.; Kirk, K.; Saliba, K.J. Coenzyme A biosynthesis: An antimicrobial drug target. FEMS Microbiol. Rev. 2008, 32, 56–106. [Google Scholar] [CrossRef] [Green Version]
- Kavermann, H.; Burns, B.P.; Angermüller, K.; Odenbreit, S.; Fischer, W.; Melchers, K.; Haas, R. Identification and Characterization of Helicobacter pylori Genes Essential for Gastric Colonization. J. Exp. Med. 2003, 197, 813–822. [Google Scholar] [CrossRef]
- Yeo, H.-J.; Savvides, S.N.; Herr, A.B.; Lanka, E.; Waksman, G. Crystal Structure of the Hexameric Traffic ATPase of the Helicobacter pylori Type IV Secretion System. Mol. Cell 2000, 6, 1461–1472. [Google Scholar] [CrossRef]
- Ansari, S.; Yamaoka, Y. Survival of Helicobacter pylori in gastric acidic territory. Helicobacter 2017, 22, e12386. [Google Scholar] [CrossRef] [PubMed]
- Backert, S.; Tegtmeyer, N.; Fischer, W. Composition, structure and function of theHelicobacter pylori cagpathogenicity island encoded type IV secretion system. Future Microbiol. 2015, 10, 955–965. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, C.; Sillén, A.; Eriksson, L.; Strand, M.-L.; Enroth, H.; Normark, S.; Falk, P.; Engstrand, L. Correlation between cag Pathogenicity Island Composition and Helicobacter pylori-Associated Gastroduodenal Disease. Infect. Immun. 2003, 71, 6573–6581. [Google Scholar] [CrossRef] [Green Version]
- Totsika, M. Benefits and Challenges of Antivirulence Antimicrobials at the Dawn of the Post-Antibiotic Era. Drug Deliv. Lett. 2016, 6, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Altieri, A.S.; Kelman, Z. DNA Sliding Clamps as Therapeutic Targets. Front. Mol. Biosci. 2018, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Verma, V.; Gautam, G.; Kumari, N.; Dhar, S.K.; Gourinath, S. Targeting the β-clamp in Helicobacter pylori with FDA-approved drugs reveals micromolar inhibition by diflunisal. FEBS Lett. 2017, 591, 2311–2322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kling, A.; Lukat, P.; Almeida, D.V.; Bauer, A.; Fontaine, E.; Sordello, S.; Zaburannyi, N.; Herrmann, J.; Wenzel, S.C.; König, C.; et al. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 2015, 348, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Zeng, J.; Xie, J. Antibiotic drugs targeting bacterial RNAs. Acta Pharm. Sin. B 2014, 4, 258–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heath, R.J.; White, W.S.; Rock, C.O. Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 2001, 40, 467–497. [Google Scholar] [CrossRef]
- Pereira, L.; Hoover, T.R. Stable Accumulation of σ54 in Helicobacter pylori Requires the Novel Protein HP0958. J. Bacteriol. 2005, 187, 4463–4469. [Google Scholar] [CrossRef] [Green Version]
- Mu, W.; Yu, S.; Zhu, L.; Zhang, T.; Jiang, B. Recent research on 3-phenyllactic acid, a broad-spectrum antimicrobial compound. Appl. Microbiol. Biotechnol. 2012, 95, 1155–1163. [Google Scholar] [CrossRef] [PubMed]
- Adamczak, A.; Ożarowski, M.; Karpiński, T.M. Antibacterial Activity of Some Flavonoids and Organic Acids Widely Distributed in Plants. J. Clin. Med. 2020, 9, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, S.; Zeng, W.; Luo, F.; Zhao, J.; Yang, Z.; Sun, Q. Antibacterial activity of organic acids in aqueous extracts from pine needles (Pinus massoniana Lamb.). Food Sci. Biotechnol. 2010, 19, 35–41. [Google Scholar] [CrossRef]
- Jadamus, A.; Vahjen, W.; Simon, O. Studies on the mode of action of probiotics: Effects of the spore-specific dipicolinic acid on selected intestinal bacteria. J. Agric. Sci. 2005, 143, 529–535. [Google Scholar] [CrossRef]
- Coban, H.B. Organic acids as antimicrobial food agents: Applications and microbial productions. Bioprocess. Biosyst. Eng. 2020, 43, 569–591. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Ren, L.; Zhou, Y.; Ye, B. Characterization of antimicrobial activity of three Lactobacillus plantarum strains isolated from Chinese traditional dairy food. Food Sci. Nutr. 2019, 7, 1997–2005. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Ricke, S.C. Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poult. Sci. 2003, 82, 632–639. [Google Scholar] [CrossRef]
- Chen, X.; Liu, X.-M.; Tian, F.; Zhang, Q.; Zhang, H.-P.; Zhang, H.; Chen, W. Antagonistic Activities of Lactobacilli against Helicobacter pylori Growth and Infection in Human Gastric Epithelial Cells. J. Food Sci. 2012, 77, M9–M14. [Google Scholar] [CrossRef]
- Lin, W.-H.; Lin, C.-K.; Sheu, S.-J.; Hwang, C.-F.; Ye, W.-T.; Hwang, W.-Z.; Tsen, H.-Y. Antagonistic Activity of Spent Culture Supernatants of Lactic Acid Bacteria against Helicobacter Pylori Growth and Infection in Human Gastric Epithelial AGS Cells. J. Food Sci. 2009, 74, M225–M230. [Google Scholar] [CrossRef]
- Matsushima, M.; Suzuki, T.; Masui, A.; Kasai, K.; Kouchi, T.; Takagi, A.; Shirai, T.; Mine, T. Growth inhibitory action of cranberry onHelicobacter pylori. J. Gastroenterol. Hepatol. 2008, 23, S175–S180. [Google Scholar] [CrossRef] [PubMed]
- Na, J.-H.; An, Y.J.; Cha, S.-S. GMP and IMP Are Competitive Inhibitors of CMY-10, an Extended-Spectrum Class C β-Lactamase. Antimicrob. Agents Chemother. 2017, 61, e00098-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Na, J.-H.; Lee, T.H.; Park, S.-B.; Kim, M.-K.; Jeong, B.-G.; Chung, K.M.; Cha, S.-S. In vitro and in vivo Inhibitory Activity of NADPH Against the AmpC BER Class C β-Lactamase. Front. Cell. Infect. Microbiol. 2018, 8, 441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, J.; Wu, Y.; Zhong, K.; Xiao, K.; Liu, L.; Huang, Y.; Wang, Z.; Gao, H. A Comparative Study on the Effects of Quinic Acid and Shikimic Acid on Cellular Functions of Staphylococcus aureus. J. Food Prot. 2018, 81, 1187–1192. [Google Scholar] [CrossRef]
- Mehla, K.; Ramana, J. Novel Drug Targets for Food-Borne Pathogen Campylobacter jejuni: An Integrated Subtractive Genomics and Comparative Metabolic Pathway Study. OMICS 2015, 19, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Zazgornik, J.; Mittermayer, H. Citric acid inhibits growth of Helicobacter pylori in vitro: A new strategy for eradication. Wien. Klin. Wochenschr. 2011, 123, 38–40. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Luo, H.; Lin, Y.; Gao, F.; Zhang, Z.; Zhang, R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014, 42, D574–D580. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Ou, H.; Zhang, C. DEG: A database of essential genes. Nucleic Acids Res. 2004, 32, D271–D272. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Piccolomini, R.; Di Bonaventura, G.; Catamo, G.; Carbone, F.; Neri, M. Comparative evaluation of the E test, agar dilution, and broth microdilution for testing susceptibilities of Helicobacter pylori strains to 20 antimicrobial agents. J. Clin. Microbiol. 1997, 35, 1842–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; CLSI Guidline M45; CLSI: Wayne, PA, USA, 2015. [Google Scholar]

| Target | Protein Name | App | Pathway | Similarity to Common Pathogen | Possible Ligands | Ligand Drugbank Accession Number |
|---|---|---|---|---|---|---|
| AroE | Shikimate dehydrogenase | PW EP | Phenylalanine, tyrosine, and tryptophan biosynthesis | Mostly 80–200 | 1,4-Dithiothreitol | DB04447 |
| Nicotinamide adenine dinucleotide phosphate | DB03461 | |||||
| 2′-Monophosphoadenosine-5′-diphosphate | DB02363 | |||||
| AroQ | 3-dehydroquinate dehydratase | CP PW EP | Phenylalanine, tyrosine, and tryptophan biosynthesis | Mostly 80–200 | 3-Dehydroquinic Acid | DB03868 |
| N-(1,4-dihydro-5H-tetrazol-5-ylidene)-9-oxo-9H-xanthene-2-sulfonamide | DB04698 | |||||
| 2,3-Anhydro-quinic acid | DB02801 | |||||
| 3-Hydroxyimino quinic acid | DB03739 | |||||
| 2-Anhydro-3-fluoro-quinic acid | DB02786 | |||||
| 3-Dehydroshikimate | DB04347 | |||||
| 1,3,4-Trihydroxy-5-(3-phenoxypropyl)-cyclohexane-1-carboxylic acid | DB04656 | |||||
| (1s,4s,5s)-1,4,5-Trihydroxy-3-[3-(phenylthio)phenyl]cyclohex-2-ene-1-carboxylic acid | DB08485 | |||||
| CheY | chemotaxis protein | PW EP | Two-component system and bacterial chemotaxis | >200 | S-Methyl Phosphocysteine | DB02461 |
| (S)-Aspartimide | DB03487 | |||||
| Aspartate Beryllium Trifluoride | DB04156 | |||||
| Adenosine-5′-Rp-Alpha-Thio-Triphosphate | DB02355 | |||||
| alpha,beta-Methyleneadenosine 5′-triphosphate | DB02596 | |||||
| 2-Hydroxyestradiol | DB07706 | |||||
| Guanosine-5′-Monophosphate | DB01972 | |||||
| Phosphoaspartate | DB01857 | |||||
| CmoA | Carboxy-S-adenosyl-L-methionine synthase | CP EP | 5-(methoxycarbonylmethoxy)uridine biosynthesis | >200 | S-Adenosyl-L-Homoselenocysteine | DB03423 |
| DapB | 4-hydroxy-tetrahydrodipicolinate reductase | CP PW EP | Lysine biosynthesis | >200 | 3-Acetylpyridine Adenine Dinucleotide | DB03363 |
| Dipicolinic acid | DB04267 | |||||
| Ddl | D-alanine-D-alanine ligase | CP PW EP | D-alanine metabolism and Peptidoglycan biosynthesis | >200 | 3-Chloro-2,2-dimethyl-n-[4-(trifluoromethyl)phenyl]propanamide | DB07805 |
| DnaN | DNA polymerase III subunit β | PW EP | DNA replication, Mismatch repair, and Homologous recombination | >200 | [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | DB06998 |
| HP0405 | Hypothetical protein 0405 | EP | NA | >200 | Selenocysteine | DB02345 |
| S-Mercaptocysteine | DB02761 | |||||
| S-Selanyl Cysteine | DB03049 | |||||
| L-2-amino-3-butynoic acid | DB04217 | |||||
| 3′-O-N-Octanoyl-a-D-Glucopyranosyl-B-D-Fructofuranoside | DB02346 | |||||
| IspA | Geranyl diphosphate synthase | CP PW EP | Terpenoid backbone biosynthesis | >200 | Pyrophosphoric acid | DB04160 |
| Isopentyl Pyrophosphate | DB02508 | |||||
| Dimethylallyl S-Thiolodiphosphate | DB02270 | |||||
| LacA/ Rpi | Ribose-5-phosphate isomerase B | PW EP | Pentose phosphate pathway, Fructose and mannose metabolism | 80–200 | 2′-Monophosphoadenosine-5′-Diphosphate | DB02363 |
| LpxA | Acyl-[acyl-carrier-protein]—UDP-N-acetylglucosamine O-acyltransferase | PW EP | Lipopolysaccharide biosynthesis | >200 | D-tartaric acid | DB01694 |
| 2-Hydroxymethyl-6-octylsulfanyl-tetrahydro-pyran-3,4,5-triol | DB08558 | |||||
| 4-Chloro-N-(3-methoxypropyl)-N-[(3S)-1-(2-phenylethyl)piperidin-3-yl]benzamide | DB08344 | |||||
| PanD | Aspartate 1-decarboxylase proenzyme | PW EP | β-alanine metabolism | 80–200 | Malonic acid | DB02175 |
| S-oxy-L-cysteine | DB03382 | |||||
| RibC | Riboflavin synthase | CP PW EP | Riboflavin metabolism | >200 | Citric acid | DB04272 |
| Flavin mononucleotide | DB03247 | |||||
| Lumichrome | DB04345 | |||||
| RplE | 50S ribosomal subunit L5 | PW EP | Ribosome | >200 | (S)-3-phenyllactic acid | DB02494 |
| RpsF | 30S ribosomal subunit S6 | PW EP | Ribosome | 80–200 | 2-Methylthio-n6-isopentenyl-adenosine-5′-monophosphate | DB08185 |
| RpsJ | 30S ribosomal subunit S10 | PW EP | Ribosome | 80–200 | 2-Methylthio-n6-isopentenyl-adenosine-5′-monophosphate | DB08185 |
| TrbB/VirB11_2 | Type IV secretion system ATPase | PW EP | Bacterial secretion system Epithelial cell signaling | Mostly 80–200 | 2′-Monophosphoadenosine-5′-Diphosphate | DB02363 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim, K.A.; Helmy, O.M.; Kashef, M.T.; Elkhamissy, T.R.; Ramadan, M.A. Identification of Potential Drug Targets in Helicobacter pylori Using In Silico Subtractive Proteomics Approaches and Their Possible Inhibition through Drug Repurposing. Pathogens 2020, 9, 747. https://doi.org/10.3390/pathogens9090747
Ibrahim KA, Helmy OM, Kashef MT, Elkhamissy TR, Ramadan MA. Identification of Potential Drug Targets in Helicobacter pylori Using In Silico Subtractive Proteomics Approaches and Their Possible Inhibition through Drug Repurposing. Pathogens. 2020; 9(9):747. https://doi.org/10.3390/pathogens9090747
Chicago/Turabian StyleIbrahim, Kareem A., Omneya M. Helmy, Mona T. Kashef, Tharwat R. Elkhamissy, and Mohammed A. Ramadan. 2020. "Identification of Potential Drug Targets in Helicobacter pylori Using In Silico Subtractive Proteomics Approaches and Their Possible Inhibition through Drug Repurposing" Pathogens 9, no. 9: 747. https://doi.org/10.3390/pathogens9090747





