Biointerface Materials for Cellular Adhesion: Recent Progress and Future Prospects
Abstract
1. Introduction
2. Biointerfaces for Non-Specific Cell Type Adhesion
2.1. Current Trends in Biointerface Materials for Non-Specific Cell Type Adhesion
2.1.1. Utilization of Multiple Materials and Processes for Novel Biointerface Material Properties
2.1.2. Biointerface Materials with Integrated ECM and ECM-Related Elements
2.1.3. Biointerface Materials for Three-Dimensional Applications
| Method | Base Scaffold Material | Scaffold Shape | Cell Type | Described 1 Potential Applications | Ref. |
|---|---|---|---|---|---|
| Molded | Hydrogel (cellulose) | Custom | Mouse fibroblast | Drug delivery and tissue engineering | [75] |
| 3D printed | Hydrogel (polyHIPEs) | Custom | Human MSC | Synthetic bone graft and tissue engineering | [76] |
| Electrospun | Hydrogel (GelMA) | Thread | Human fibroblast | Synthetic skin graft, tissue engineering and wound dressing | [77] |
| Hanging droplet | Hydrogel (alginate) | Spherical | Human colon cells and human liver cells | Drug screening | [78] |
| Decellularized tissue | ECM | Originating tissue | Human liver cells | Artificial organ, disease modelling, drug screening and tissue engineering | [79] |
| Direct laser writing | Protein-functionalized photoresist | Custom | Human lung adenocarcenoma and mouse embryo fibroblast | Tissue engineering and regenerative medicine | [80] |
3. Biointerfaces for Cell Type-Specific Adhesion
3.1. Current Trends in Biointerface Materials for Cell Type-Specific Adhesion
3.1.1. Precise and Portable Medical Diagnostics
3.1.2. Biocompatible and Implantable Materials
4. Considerations for Selecting Biointerface Materials for Cell Adhesion
4.1. Dimensions of Cell Occupation
4.2. Target Cell(s) of Interest
4.3. Application-Specific Challenges
4.4. Cell Detection Methods
5. Dynamic and Smart Materials for Biological Applications
6. Summary and Conclusions
7. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Huang, S.; Ingber, D.E. The structural and mechanical complexity of cell-growth control. Nat. Cell Biol. 1999, 1, E131–E138. [Google Scholar] [CrossRef]
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular Matrix degradation and remodeling in development and disease. Cold Spring Harbor Perspect. Biol. 2011, 3, a005058. [Google Scholar] [CrossRef]
- Nitsan, I.; Drori, S.; Lewis, Y.E.; Cohen, S.; Tzlil, S. Mechanical communication in cardiac cell synchronized beating. Nat. Phys. 2016, 12, 472–477. [Google Scholar] [CrossRef]
- Wang, N. Review of cellular mechanotransduction. J. Phys. D Appl. Phys. 2017, 50, 233002. [Google Scholar] [CrossRef]
- Fitzgerald, J.B.; Jin, M.; Dean, D.; Wood, D.J.; Zheng, M.H.; Grodzinsky, A.J. Mechanical Compression of Cartilage Explants Induces Multiple Time-dependent Gene Expression Patterns and Involves Intracellular Calcium and Cyclic AMP. J. Biol. Chem. 2004, 279, 19502–19511. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.B.; Jin, M.; Grodzinsky, A.J. Shear and compression differentially regulate clusters of functionally related temporal transcription patterns in cartilage tissue. J. Biol. Chem. 2006, 281, 24095–24103. [Google Scholar] [CrossRef] [PubMed]
- Ikada, Y. Surface modification of polymers for medical applications. Biomaterials 1994, 15, 725–736. [Google Scholar] [CrossRef]
- Bacakova, L.; Filova, E.; Parizek, M.; Ruml, T.; Svorcik, V. Modulation of cell adhesion, proliferation and differentiation on materials designed for body implants. Biotechnol. Adv. 2011, 29, 739–767. [Google Scholar] [CrossRef]
- Pelham, R.J.; Wang, Y.L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA 1997, 94, 13661–13665. [Google Scholar] [CrossRef]
- Wong, J.Y.; Velasco, A.; Rajagopalan, P.; Pham, Q. Directed movement of vascular smooth muscle cells on gradient-compliant hydrogels. Langmuir 2003, 19, 1908–1913. [Google Scholar] [CrossRef]
- Engler, A.; Bacakova, L.; Newman, C.; Hategan, A.; Griffin, M.; Discher, D. Substrate Compliance versus Ligand Density in Cell on Gel Responses. Biophys. J. 2004, 86, 617–628. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: A family of cell surface receptors. Cell 1987, 48, 549–554. [Google Scholar] [CrossRef]
- Gehlsen, K.R.; Dillner, L.; Engvall, E.; Ruoslahti, E. The human laminin receptor is a member of the integrin family of cell adhesion receptors. Science 1988, 241, 1228–1229. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E.; Pierschbacher, M.D. New perspectives in cell adhesion: RGD and integrins. Science 1987, 238, 491–497. [Google Scholar] [CrossRef]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 1984, 309, 30–33. [Google Scholar] [CrossRef]
- Langer, R.; Cima, L.G.; Tamada, J.A.; Wintermantel, E. Future directions in biomaterials. Biomaterials 1990, 11, 738–745. [Google Scholar] [CrossRef]
- Rowley, J.A.; Madlambayan, G.; Mooney, D.J. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials 1999, 20, 45–53. [Google Scholar] [CrossRef]
- Lévesque, S.G.; Shoichet, M.S. Synthesis of cell-adhesive dextran hydrogels and macroporous scaffolds. Biomaterials 2006, 27, 5277–5285. [Google Scholar] [CrossRef]
- Ouyang, L.; Highley, C.B.; Rodell, C.B.; Sun, W.; Burdick, J.A. 3D Printing of Shear-Thinning Hyaluronic Acid Hydrogels with Secondary Cross-Linking. ACS Biomater. Sci. Eng. 2016, 2, 1743–1751. [Google Scholar] [CrossRef]
- Qayyum, A.S.; Jain, E.; Kolar, G.; Kim, Y.; Sell, S.A.; Zustiak, S.P. Design of electrohydrodynamic sprayed polyethylene glycol hydrogel microspheres for cell encapsulation. Biofabrication 2017, 9. [Google Scholar] [CrossRef]
- Huang, H.; Lovell, J.F. Advanced Functional Nanomaterials for Theranostics. Adv. Funct. Mater. 2017, 27. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Liu, X.; Wang, G.; Li, M.; Bratlie, K.M.; Cochran, E.; Wang, Q. Polymeric multifunctional nanomaterials for theranostics. J. Mater. Chem. B 2015, 3, 6856–6870. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.K.; Kim, T.; Paik, S.; Haam, S.; Huh, Y.M.; Lee, K. Nanomaterials for theranostics: Recent advances and future challenges. Chem. Rev. 2015, 115, 327–394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, Q. Biosensors and bioelectronics on smartphone for portable biochemical detection. Biosens. Bioelectron. 2016, 75, 273–284. [Google Scholar] [CrossRef]
- Harrison, R.G.; Greenman, M.J.; Mall, F.P.; Jackson, C.M. Observations of the living developing nerve fiber. Anat. Rec. 1907, 1, 116–128. [Google Scholar] [CrossRef]
- Harrison, R.G. On the stereotropism of embryonic cells. Science 1911, 34, 279–281. [Google Scholar] [CrossRef]
- Curtis, A.S.G.; Forrester, J.V.; McInnes, C.; Lawrie, F. Adhesion of cells to polystyrene surfaces. J. Cell Biol. 1983, 97, 1500–1506. [Google Scholar] [CrossRef]
- Martin, G.R.; Rubin, H. Effects of cell adhesion to the substratum on the growth of chick embryo fibroblasts. Exp. Cell Res. 1974, 85, 319–333. [Google Scholar] [CrossRef]
- Bentley, K.L.; Klebe, R.J. Fibronectin binding properties of bacteriologic petri plates and tissue culture dishes. J. Biomed. Mater. Res. 1985, 19, 757–769. [Google Scholar] [CrossRef]
- Ertel, S.I.; Chilkoti, A.; Horbett, T.A.; Ratner, B.D. Endothelial cell growth on oxygen-containing films deposited by radio-frequency plasmas: The role of surface carbonyl groups. J. Biomater. Sci., Polym. Ed. 1992, 3, 163–183. [Google Scholar] [CrossRef]
- Amstein, C.F.; Hartman, P.A. Adaptation of plastic surfaces for tissue culture by glow discharge. J. Clin. Microbiol. 1975, 2, 46–54. [Google Scholar] [PubMed]
- Liu, Z.Q.; Wei, Z.; Zhu, X.L.; Huang, G.Y.; Xu, F.; Yang, J.H.; Osada, Y.; Zrínyi, M.; Li, J.H.; Chen, Y.M. Dextran-based hydrogel formed by thiol-Michael addition reaction for 3D cell encapsulation. Colloids Surf. B 2015, 128, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Utech, S.; Prodanovic, R.; Mao, A.S.; Ostafe, R.; Mooney, D.J.; Weitz, D.A. Microfluidic Generation of Monodisperse, Structurally Homogeneous Alginate Microgels for Cell Encapsulation and 3D Cell Culture. Adv. Healthc. Mater. 2015, 4, 1628–1633. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Yu, S.; Liu, B.; Ni, Y.; Yu, C.; Su, Y.; Zhu, X.; Yu, X.; Zhou, Y.; Yan, D. An Injectable Enzymatically Crosslinked Carboxymethylated Pullulan/Chondroitin Sulfate Hydrogel for Cartilage Tissue Engineering. Sci. Rep. 2016, 6, 20014. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Guo, J.; Tierney, E.G.; Curtin, C.M.; Malhotra, M.; Darcy, R.; O’Brien, F.J.; O’Driscoll, C.M. The use of collagen-based scaffolds to simulate prostate cancer bone metastases with potential for evaluating delivery of nanoparticulate gene therapeutics. Biomaterials 2015, 66, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Ceseracciu, L.; Heredia-Guerrero, J.A.; Dante, S.; Athanassiou, A.; Bayer, I.S. Robust and biodegradable elastomers based on corn starch and polydimethylsiloxane (PDMS). ACS Appl. Mater. Interfaces 2015, 7, 3742–3753. [Google Scholar] [CrossRef] [PubMed]
- Giobbe, G.G.; Crowley, C.; Luni, C.; Campinoti, S.; Khedr, M.; Kretzschmar, K.; de Santis, M.M.; Zambaiti, E.; Michielin, F.; Meran, L.; et al. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat. Commun. 2019, 10, 5658. [Google Scholar] [CrossRef]
- Nguyen, T.-U.; Bashur, C.A.; Kishore, V. Impact of elastin incorporation into electrochemically aligned collagen fibers on mechanical properties and smooth muscle cell phenotype. Biomed. Mater. 2016, 11, 025008. [Google Scholar] [CrossRef]
- Sireesha, M.; Jagadeesh Babu, V.; Ramakrishna, S. Biocompatible and biodegradable elastomer/fibrinogen composite electrospun scaffolds for cardiac tissue regeneration. RSC Adv. 2015, 5, 103308–103314. [Google Scholar] [CrossRef]
- Hsiao, C.-T.; Cheng, H.W.; Huang, C.M.; Li, H.R.; Ou, M.H.; Huang, J.R.; Khoo, K.H.; Yu, H.W.; Chen, Y.Q.; Wang, Y.K.; et al. Fibronectin in cell adhesion and migration via N-glycosylation. Oncotarget 2017, 8, 70653–70668. [Google Scholar] [CrossRef]
- Li, L.; Lu, C.; Wang, L.; Chen, M.; White, J.; Hao, X.; McLean, K.M.; Chen, H.; Hughes, T.C. Gelatin-Based Photocurable Hydrogels for Corneal Wound Repair. ACS Appl. Mater. Interfaces 2018, 10, 13283–13292. [Google Scholar] [CrossRef] [PubMed]
- Hyysalo, A.; Ristola, M.; Mäkinen, M.E.L.; Häyrynen, S.; Nykter, M.; Narkilahti, S. Laminin α5 substrates promote survival, network formation and functional development of human pluripotent stem cell-derived neurons in vitro. Stem Cell Res. 2017, 24, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Dolega, M.E.; Abeille, F.; Picollet-D’hahan, N.; Gidrol, X. Controlled 3D culture in Matrigel microbeads to analyze clonal acinar development. Biomaterials 2015, 52, 347–357. [Google Scholar] [CrossRef]
- Bu, X.; Li, J.; Yang, S.; Sun, J.; Deng, Y.; Yang, Y.; Wang, G.; Peng, Z.; He, P.; Wang, X.; et al. Surface Modification of C3N4 through Oxygen-Plasma Treatment: A Simple Way toward Excellent Hydrophilicity. ACS Appl. Mater. Interfaces 2016, 8, 31419–31425. [Google Scholar] [CrossRef] [PubMed]
- Kamande, J.W.; Nagendran, T.; Harris, J.; Taylor, A.M. Multi-compartment Microfluidic Device Geometry and Covalently Bound Poly-D-Lysine Influence Neuronal Maturation. Front. Bioeng. Biotechnol. 2019, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.; Clark, E.C.; Fong, E.L.S.; Lee, E.J.; Lu, S.; Tabata, Y.; Mikos, A.G. Evaluation of cell-laden polyelectrolyte hydrogels incorporating poly(L-Lysine) for applications in cartilage tissue engineering. Biomaterials 2016, 83, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Macková, H.; Plichta, Z.; Hlídková, H.; Sedláček, O.; Konefal, R.; Sadakbayeva, Z.; Dušková-Smrčková, M.; Horák, D.; Kubinová, Š. Reductively Degradable Poly(2-hydroxyethyl methacrylate) Hydrogels with Oriented Porosity for Tissue Engineering Applications. ACS Appl. Mater. Interfaces 2017, 9, 10544–10553. [Google Scholar] [CrossRef]
- Sanzari, I.; Buratti, E.; Huang, R.; Tusan, C.G.; Dinelli, F.; Evans, N.D.; Prodromakis, T.; Bertoldo, M. Poly(N-isopropylacrylamide) based thin microgel films for use in cell culture applications. Sci. Rep. 2020, 10, 6162. [Google Scholar] [CrossRef]
- Enayati, M.S.; Behzad, T.; Sajkiewicz, P.; Rafienia, M.; Bagheri, R.; Ghasemi-Mobarakeh, L.; Kolbuk, D.; Pahlevanneshan, Z.; Bonakdar, S.H. Development of electrospun poly (vinyl alcohol)-based bionanocomposite scaffolds for bone tissue engineering. J. Biomed. Mater. Res. Part A 2018, 106, 1111–1120. [Google Scholar] [CrossRef]
- Ahadian, S.; Ramón-Azcón, J.; Estili, M.; Liang, X.; Ostrovidov, S.; Shiku, H.; Ramalingam, M.; Nakajima, K.; Sakka, Y.; Bae, H.; et al. Hybrid hydrogels containing vertically aligned carbon nanotubes with anisotropic electrical conductivity for muscle myofiber fabrication. Sci. Rep. 2014, 4, 4271. [Google Scholar] [CrossRef]
- Zhao, W.; Odelius, K.; Edlund, U.; Zhao, C.; Albertsson, A.C. In Situ Synthesis of Magnetic Field-Responsive Hemicellulose Hydrogels for Drug Delivery. Biomacromolecules 2015, 16, 2522–2528. [Google Scholar] [CrossRef] [PubMed]
- Epstein, A.K.; Hong, D.; Kim, P.; Aizenberg, J. Biofilm attachment reduction on bioinspired, dynamic, micro-wrinkling surfaces. New J. Phys. 2013, 15. [Google Scholar] [CrossRef]
- Han, D.; Lu, Z.; Chester, S.A.; Lee, H. Micro 3D Printing of a Temperature-Responsive Hydrogel Using Projection Micro-Stereolithography. Sci. Rep. 2018, 8, 1963. [Google Scholar] [CrossRef] [PubMed]
- GhavamiNejad, A.; Samarikhalaj, M.; Aguilar, L.E.; Park, C.H.; Kim, C.S. pH/NIR light-controlled multidrug release via a mussel-inspired Nanocomposite Hydrogel for Chemo-Photothermal cancer therapy. Sci. Rep. 2016, 6, 33594. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, T.W.; Sellaro, T.L.; Badylak, S.F. Decellularization of tissues and organs. Biomaterials 2006, 27, 3675–3683. [Google Scholar] [CrossRef] [PubMed]
- Wade, R.J.; Burdick, J.A. Engineering ECM signals into biomaterials. Mater. Today 2012, 15, 454–459. [Google Scholar] [CrossRef]
- Hinderer, S.; Layland, S.L.; Schenke-Layland, K. ECM and ECM-like materials—Biomaterials for applications in regenerative medicine and cancer therapy. Adv. Drug Delivery Rev. 2016, 97, 260–269. [Google Scholar] [CrossRef]
- Mao, A.S.; Mooney, D.J. Regenerative medicine: Current therapies and future directions. Proc. Natl. Acad. Sci. USA 2015, 112, 14452–14459. [Google Scholar] [CrossRef]
- Youngblood, R.L.; Truong, N.F.; Segura, T.; Shea, L.D. It’s All in the Delivery: Designing Hydrogels for Cell and Non-viral Gene Therapies. Mol. Ther. 2018, 26, 2087–2106. [Google Scholar] [CrossRef]
- Bishop, E.S.; Mostafa, S.; Pakvasa, M.; Luu, H.H.; Lee, M.J.; Wolf, J.M.; Ameer, G.A.; He, T.C.; Reid, R.R. 3-D bioprinting technologies in tissue engineering and regenerative medicine: Current and future trends. Genes Dis. 2017, 4, 185–195. [Google Scholar] [CrossRef]
- Hippler, M.; Lemma, E.D.; Bertels, S.; Blasco, E.; Barner-Kowollik, C.; Wegener, M.; Bastmeyer, M. 3D Scaffolds to Study Basic Cell Biology. Adv. Mater. 2019, 31, 1808110. [Google Scholar] [CrossRef] [PubMed]
- Rajzer, I.; Kurowska, A.; Jabłoński, A.; Jatteau, S.; Śliwka, M.; Ziąbka, M.; Menaszek, E. Layered gelatin/PLLA scaffolds fabricated by electrospinning and 3D printing- for nasal cartilages and subchondral bone reconstruction. Mater. Des. 2018, 155, 297–306. [Google Scholar] [CrossRef]
- Badylak, S.F.; Taylor, D.; Uygun, K. Whole-Organ Tissue Engineering: Decellularization and Recellularization of Three-Dimensional Matrix Scaffolds. Annu. Rev. Biomed. Eng. 2011, 13, 27–53. [Google Scholar] [CrossRef] [PubMed]
- Yoeruek, E.; Bayyoud, T.; Maurus, C.; Hofmann, J.; Spitzer, M.S.; Bartz-Schmidt, K.-U.; Szurman, P. Decellularization of porcine corneas and repopulation with human corneal cells for tissue-engineered xenografts. Acta Ophthalmol. 2012, 90, e125–e131. [Google Scholar] [CrossRef]
- Barakat, O.; Abbasi, S.; Rodriguez, G.; Rios, J.; Wood, R.P.; Ozaki, C.; Holley, L.S.; Gauthier, P.K. Use of decellularized porcine liver for engineering humanized liver organ. J. Surg. Res. 2012, 173, e11–e25. [Google Scholar] [CrossRef]
- Abraham, S.; Sheridan, S.D.; Miller, B.; Rao, R.R. Stable propagation of human embryonic and induced pluripotent stem cells on decellularized human substrates. Biotechnol. Progr. 2010, 26, 1126–1134. [Google Scholar] [CrossRef]
- Gilpin, S.E.; Ren, X.; Okamoto, T.; Guyette, J.P.; Mou, H.; Rajagopal, J.; Mathisen, D.J.; Vacanti, J.P.; Ott, H.C. Enhanced lung epithelial specification of human induced pluripotent stem cells on decellularized lung matrix. Ann. Thorac. Surg. 2014, 98, 1721–1729. [Google Scholar] [CrossRef]
- Minami, T.; Ishii, T.; Yasuchika, K.; Fukumitsu, K.; Ogiso, S.; Miyauchi, Y.; Kojima, H.; Kawai, T.; Yamaoka, R.; Oshima, Y.; et al. Novel hybrid three-dimensional artificial liver using human induced pluripotent stem cells and a rat decellularized liver scaffold. Regen. Ther. 2019, 10, 127–133. [Google Scholar] [CrossRef]
- Kitano, K.; Schwartz, D.M.; Zhou, H.; Gilpin, S.E.; Wojtkiewicz, G.R.; Ren, X.; Sommer, C.A.; Capilla, A.V.; Mathisen, D.J.; Goldstein, A.M.; et al. Bioengineering of functional human induced pluripotent stem cell-derived intestinal grafts. Nat. Commun. 2017, 8, 765. [Google Scholar] [CrossRef]
- Lu, T.Y.; Lin, B.; Kim, J.; Sullivan, M.; Tobita, K.; Salama, G.; Yang, L. Repopulation of decellularized mouse heart with human induced pluripotent stem cell-derived cardiovascular progenitor cells. Nat. Commun. 2013, 4, 2307. [Google Scholar] [CrossRef]
- Yu, L.; Hou, Y.; Xie, W.; Camacho, J.L.C.; Cheng, C.; Holle, A.; Young, J.; Trappmann, B.; Zhao, W.; Melzig, M.F.; et al. Ligand Diffusion Enables Force-Independent Cell Adhesion via Activating α5β1 Integrin and Initiating Rac and RhoA Signaling. Adv. Mater. 2020, 32, 2002566. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Zhao, C.; Spatz, J.P.; Wei, Q. Nanopatterned Adhesive, Stretchable Hydrogel to Control Ligand Spacing and Regulate Cell Spreading and Migration. ACS Nano 2017, 11, 8282–8291. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Yu, L.; Xie, W.; Camacho, L.C.; Zhang, M.; Chu, Z.; Wei, Q.; Haag, R. Surface Roughness and Substrate Stiffness Synergize to Drive Cellular Mechanoresponse. Nano Lett. 2020, 20, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Xie, W.; Yu, L.; Camacho, L.C.; Nie, C.; Zhang, M.; Haag, R.; Wei, Q. Surface Roughness Gradients Reveal Topography-Specific Mechanosensitive Responses in Human Mesenchymal Stem Cells. Small 2020, 16, 1905422. [Google Scholar] [CrossRef]
- Yang, X.; Liu, G.; Peng, L.; Guo, J.; Tao, L.; Yuan, J.; Chang, C.; Wei, Y.; Zhang, L. Highly Efficient Self-Healable and Dual Responsive Cellulose-Based Hydrogels for Controlled Release and 3D Cell Culture. Adv. Funct. Mater. 2017, 27. [Google Scholar] [CrossRef]
- Whitely, M.; Cereceres, S.; Dhavalikar, P.; Salhadar, K.; Wilems, T.; Smith, B.; Mikos, A.; Cosgriff-Hernandez, E. Improved in situ seeding of 3D printed scaffolds using cell-releasing hydrogels. Biomaterials 2018, 185, 194–204. [Google Scholar] [CrossRef]
- Zhao, X.; Sun, X.; Yildirimer, L.; Lang, Q.; Lin, Z.Y.; Zheng, R.; Zhang, Y.; Cui, W.; Annabi, N.; Khademhosseini, A. Cell infiltrative hydrogel fibrous scaffolds for accelerated wound healing. Acta Biomater. 2017, 49, 66–77. [Google Scholar] [CrossRef]
- Aeby, E.A.; Misun, P.M.; Hierlemann, A.; Frey, O. Microfluidic Hydrogel Hanging-Drop Network for Long-Term Culturing of 3D Microtissues and Simultaneous High-Resolution Imaging. Adv. Biosyst. 2018, 2, 1800054. [Google Scholar] [CrossRef]
- Mazza, G.; Rombouts, K.; Rennie Hall, A.; Urbani, L.; Vinh Luong, T.; Al-Akkad, W.; Longato, L.; Brown, D.; Maghsoudlou, P.; Dhillon, A.P.; et al. Decellularized human liver as a natural 3D-scaffold for liver bioengineering and transplantation. Sci. Rep. 2015, 5, 13079. [Google Scholar] [CrossRef]
- Richter, B.; Hahn, V.; Bertels, S.; Claus, T.K.; Wegener, M.; Delaittre, G.; Barner-Kowollik, C.; Bastmeyer, M. Guiding Cell Attachment in 3D Microscaffolds Selectively Functionalized with Two Distinct Adhesion Proteins. Adv. Mater. 2017, 29. [Google Scholar] [CrossRef]
- Hou, H.W.; Petchakup, C.; Tay, H.M.; Tam, Z.Y.; Dalan, R.; Chew, D.E.K.; Li, H.; Boehm, B.O. Rapid and label-free microfluidic neutrophil purification and phenotyping in diabetes mellitus. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Černochová, P.; Blahová, L.; Medalová, J.; Nečas, D.; Michlíček, M.; Kaushik, P.; Přibyl, J.; Bartošíková, J.; Manakhov, A.; Bačáková, L.; et al. Cell type specific adhesion to surfaces functionalised by amine plasma polymers. Sci. Rep. 2020, 10, 9357. [Google Scholar] [CrossRef] [PubMed]
- Benvenuto, P.; Neves, M.A.D.; Blaszykowski, C.; Romaschin, A.; Chung, T.; Kim, S.R.; Thompson, M. Adlayer-mediated antibody immobilization to stainless steel for potential application to endothelial progenitor cell capture. Langmuir 2015, 31, 5423–5431. [Google Scholar] [CrossRef]
- Lin, Q.K.; Hou, Y.; Xu, X.; Tang, J.; Han, Y.; Chen, H.; Ji, J. Anti-CD34 antibody functionalized swollen polymeric coating for endothelial cell rapid selectively capture. Int. J. Polym. Mater. Polym. Biomater. 2015, 64, 99–103. [Google Scholar] [CrossRef]
- Myung, J.H.; Roengvoraphoj, M.; Tam, K.A.; Ma, T.; Memoli, V.A.; Dmitrovsky, E.; Freemantle, S.J.; Hong, S. Effective Capture of Circulating Tumor Cells from a Transgenic Mouse Lung Cancer Model Using Dendrimer Surfaces Immobilized with Anti-EGFR. Anal. Chem. 2015, 87, 10096–10102. [Google Scholar] [CrossRef]
- Kimura, T.; Nakamura, N.; Sasaki, N.; Hashimoto, Y.; Sakaguchi, S.; Kimura, S.; Kishida, A. Capture and release of target cells using a surface that immobilizes an antibody via desthiobiotin-avidin interaction. Sens. Mater. 2016, 28, 1255–1263. [Google Scholar] [CrossRef]
- Kimura, T.; Nakamura, N.; Umeda, K.; Hashimoto, Y.; Kishida, A. Capture and release of cells using a temperature-responsive surface that immobilizes an antibody through DNA duplex formation. J. Biomater. Sci., Polym. Ed. 2017, 28, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.-K.; Nikolov, P.M.; Giselbrecht, S.; Niemeyer, C.M. DNA-SMART: Biopatterned Polymer Film Microchannels for Selective Immobilization of Proteins and Cells. Small 2017, 13, 1603923. [Google Scholar] [CrossRef]
- Yan, S.; Chen, P.; Zeng, X.; Zhang, X.; Li, Y.; Xia, Y.; Wang, J.; Dai, X.; Feng, X.; Du, W.; et al. Integrated Multifunctional Electrochemistry Microchip for Highly Efficient Capture, Release, Lysis, and Analysis of Circulating Tumor Cells. Anal. Chem. 2017, 89, 12039–12044. [Google Scholar] [CrossRef]
- Qi, P.; Yan, W.; Yang, Y.; Li, Y.; Fan, Y.; Chen, J.; Yang, Z.; Tu, Q.; Huang, N. Immobilization of DNA aptamers via plasma polymerized allylamine film to construct an endothelial progenitor cell-capture surface. Colloids Surf. B 2015, 126, 70–79. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Castellanos, C.A.; King, M.R. Immobilized surfactant-nanotube complexes support selectin-mediated capture of viable circulating tumor cells in the absence of capture antibodies. J. Biomed. Mater. Res. Part A 2015, 103, 3407–3418. [Google Scholar] [CrossRef] [PubMed]
- Komoriya, A.; Green, L.J.; Mervic, M.; Yamada, S.S.; Yamada, K.M.; Humphries, M.J. The minimal essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J. Biol. Chem. 1991, 266, 15075–15079. [Google Scholar] [PubMed]
- Humphries, M.J.; Akiyama, S.K.; Komoriya, A.; Olden, K.; Yamada, K.M. Identification of an alternatively spliced site in human plasma fibronectin that mediates cell type-specific adhesion. J. Cell Biol. 1986, 103, 2637–2647. [Google Scholar] [CrossRef]
- Kimura, T.; Nakamura, N.; Hashimoto, Y.; Sakaguchi, S.; Kimura, S.; Kishida, A. Selective cell capture and release using antibody-immobilized polymer-grafted surface. Kobunshi Ronbunshu 2018, 75, 155–163. [Google Scholar] [CrossRef]
- Laksanasopin, T.; Guo, T.W.; Nayak, S.; Sridhara, A.A.; Xie, S.; Olowookere, O.O.; Cadinu, P.; Meng, F.; Chee, N.H.; Kim, J.; et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 2015, 7, 273re1. [Google Scholar] [CrossRef] [PubMed]
- Striebel, J.; Vorobii, M.; Kumar, R.; Liu, H.-Y.; Yang, B.; Weishaupt, C.; Rodriguez-Emmenegger, C.; Fuchs, H.; Hirtz, M.; Riehemann, K. Controlled Surface Adhesion of Macrophages via Patterned Antifouling Polymer Brushes. Adv. NanoBiomed Res. 2020. [Google Scholar] [CrossRef]
- Knowlton, S.; Joshi, A.; Syrrist, P.; Coskun, A.F.; Tasoglu, S. 3D-printed smartphone-based point of care tool for fluorescence- and magnetophoresis-based cytometry. Lab Chip 2017, 17, 2839–2851. [Google Scholar] [CrossRef]
- Tran, M.V.; Susumu, K.; Medintz, I.L.; Algar, W.R. Supraparticle Assemblies of Magnetic Nanoparticles and Quantum Dots for Selective Cell Isolation and Counting on a Smartphone-Based Imaging Platform. Anal. Chem. 2019, 91, 11963–11971. [Google Scholar] [CrossRef]
- Van der Toom, E.E.; Verdone, J.E.; Gorin, M.A.; Pienta, K.J. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget 2016, 7, 62754–62766. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Brinkmann, F.; Hirtz, M.; Haller, A.; Gorges, T.M.; Vellekoop, M.J.; Riethdorf, S.; Müller, V.; Pantel, K.; Fuchs, H. A versatile microarray platform for capturing rare cells. Sci. Rep. 2015, 5, 15342. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Koch, C.; Haller, A.; Joosse, S.A.; Kumar, R.; Vellekoop, M.J.; Horst, L.J.; Keller, L.; Babayan, A.; Failla, A.V.; et al. Evaluation of Microfluidic Ceiling Designs for the Capture of Circulating Tumor Cells on a Microarray Platform. Adv. Biosyst. 2020, 4, 1900162. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Donovan, M.J.; Jiang, J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013, 113, 2842–2862. [Google Scholar] [CrossRef] [PubMed]
- Jana, S. Endothelialization of cardiovascular devices. Acta Biomater. 2019, 99, 53–71. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Breslin, S.; O’Driscoll, L. The relevance of using 3D cell cultures, in addition to 2D monolayer cultures, when evaluating breast cancer drug sensitivity and resistance. Oncotarget 2016, 7, 45745–45756. [Google Scholar] [CrossRef]
- Tsuyuki, E.; Tsuyuki, H.; Stahmann, M.A. The synthesis and enzymatic hydrolysis of poly-D-lysine. J. Biol. Chem. 1956, 222, 479–485. [Google Scholar]
- Poldervaart, M.T.; Gremmels, H.; van Deventer, K.; Fledderus, J.O.; Öner, F.C.; Verhaar, M.C.; Dhert, W.J.A.; Alblas, J. Prolonged presence of VEGF promotes vascularization in 3D bioprinted scaffolds with defined architecture. J. Controlled Release 2014, 184, 58–66. [Google Scholar] [CrossRef]
- Cavo, M.; Caria, M.; Pulsoni, I.; Beltrame, F.; Fato, M.; Scaglione, S. A new cell-laden 3D Alginate-Matrigel hydrogel resembles human breast cancer cell malignant morphology, spread and invasion capability observed “in vivo”. Sci. Rep. 2018, 8, 5333. [Google Scholar] [CrossRef] [PubMed]
- Nabovati, G.; Ghafar-Zadeh, E.; Letourneau, A.; Sawan, M. Smart Cell Culture Monitoring and Drug Test Platform Using CMOS Capacitive Sensor Array. IEEE Trans. Biomed. Eng. 2019, 66, 1094–1104. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Wen, X.; Tan, X.H.M.; Chiou, P.Y. Plasmonic micropillars for precision cell force measurement across a large field-of-view. Appl. Phys. Lett. 2018, 112, 033701. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Dauriac, V.; Thibert, V.; Senechal, H.; Peltre, G.; Zhang, X.X.; Descroix, S. Micropillar array chips toward new immunodiagnosis. Lab Chip 2010, 10, 2597–2604. [Google Scholar] [CrossRef] [PubMed]
- Ghafar-Zadeh, E.; Sawan, M. CMOS Capacitive Sensors for Lab-on-Chip Applications; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar] [CrossRef]
- Ghafar-Zadeh, E.; Waldeisen, J.R.; Lee, L.P. Engineered approaches to the stem cell microenvironment for cardiac tissue regeneration. Lab Chip 2011, 11, 3031–3048. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.S.; Simões, B.; Mendes, P.M. The increasing dynamic, functional complexity of bio-interface materials. Nat. Rev. Chem. 2018, 2. [Google Scholar] [CrossRef]
- Hippler, M.; Blasco, E.; Qu, J.; Tanaka, M.; Barner-Kowollik, C.; Wegener, M.; Bastmeyer, M. Controlling the shape of 3D microstructures by temperature and light. Nat. Commun. 2019. [Google Scholar] [CrossRef]
- Guo, Z.; Liu, H.; Dai, W.; Lei, Y. Responsive principles and applications of smart materials in biosensing. Smart Mater. Med. 2020, 1, 54–65. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, J.; Song, J.; Yang, J.; Du, Z.; Zhao, W.; Guo, H.; Wen, C.; Li, Q.; Sui, X.; et al. A Multifunctional Pro-Healing Zwitterionic Hydrogel for Simultaneous Optical Monitoring of pH and Glucose in Diabetic Wound Treatment. Adv. Funct. Mater. 2020, 30, 1905493. [Google Scholar] [CrossRef]
- Duan, D.; Fan, K.; Zhang, D.; Tan, S.; Liang, M.; Liu, Y.; Zhang, J.; Zhang, P.; Liu, W.; Qiu, X.; et al. Nanozyme-strip for rapid local diagnosis of Ebola. Biosens. Bioelectron. 2015, 74, 134–141. [Google Scholar] [CrossRef]
- Parlak, O.; Beyazit, S.; Tse-Sum-Bui, B.; Haupt, K.; Turner, A.P.F.; Tiwari, A. Programmable bioelectronics in a stimuli-encoded 3D graphene interface. Nanoscale 2016, 8, 9976–9981. [Google Scholar] [CrossRef] [PubMed]
- Balán, I.C.; Lopez-Rios, J.; Nayak, S.; Lentz, C.; Arumugam, S.; Kutner, B.; Dolezal, C.; Macar, O.U.; Pabari, T.; Wang Ying, A.; et al. SMARTtest: A Smartphone App to Facilitate HIV and Syphilis Self- and Partner-Testing, Interpretation of Results, and Linkage to Care. AIDS Behav. 2020, 24, 1560–1573. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Geiger, H. HSC Niche Biology and HSC Expansion Ex Vivo. Trends Mol. Med. 2017, 23, 799–819. [Google Scholar] [CrossRef] [PubMed]
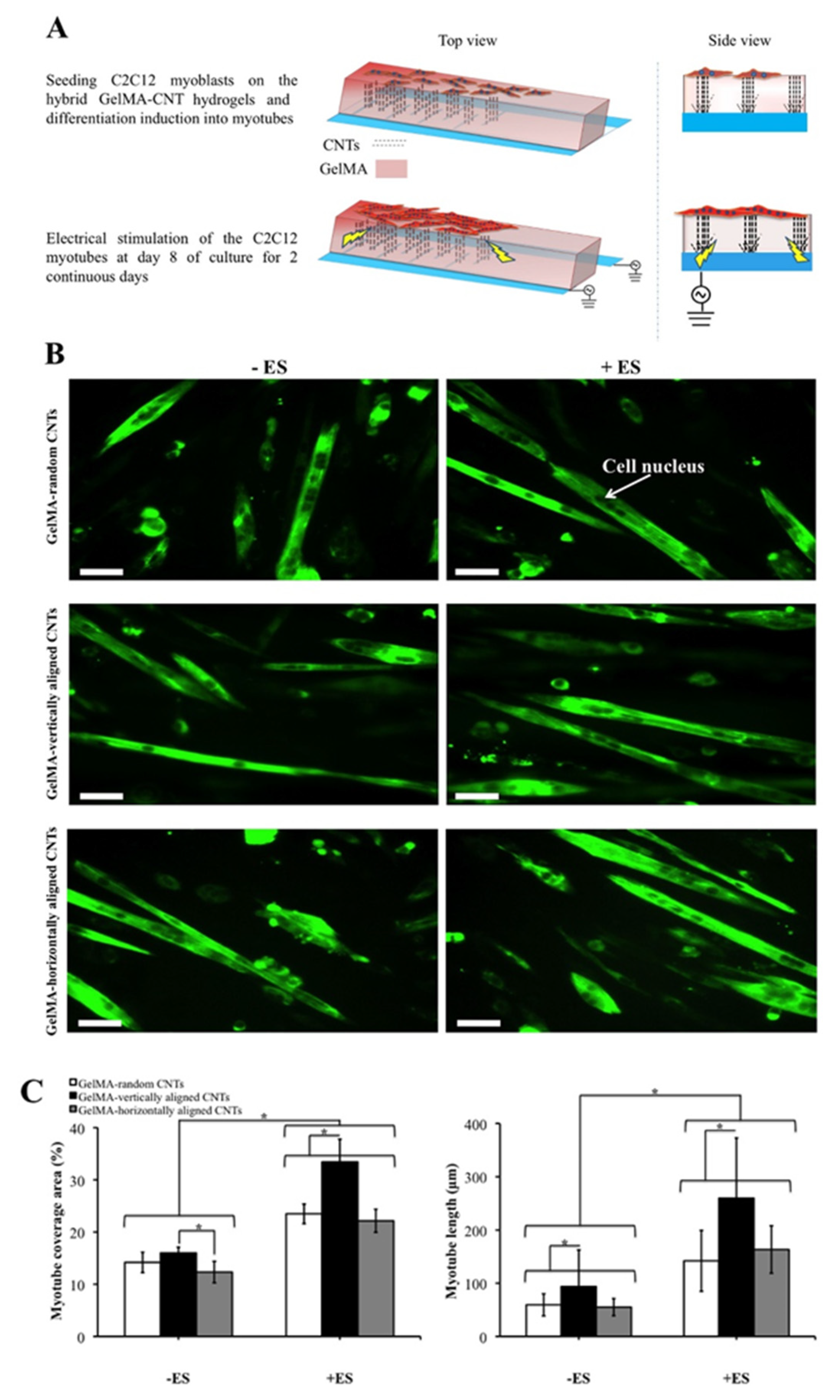
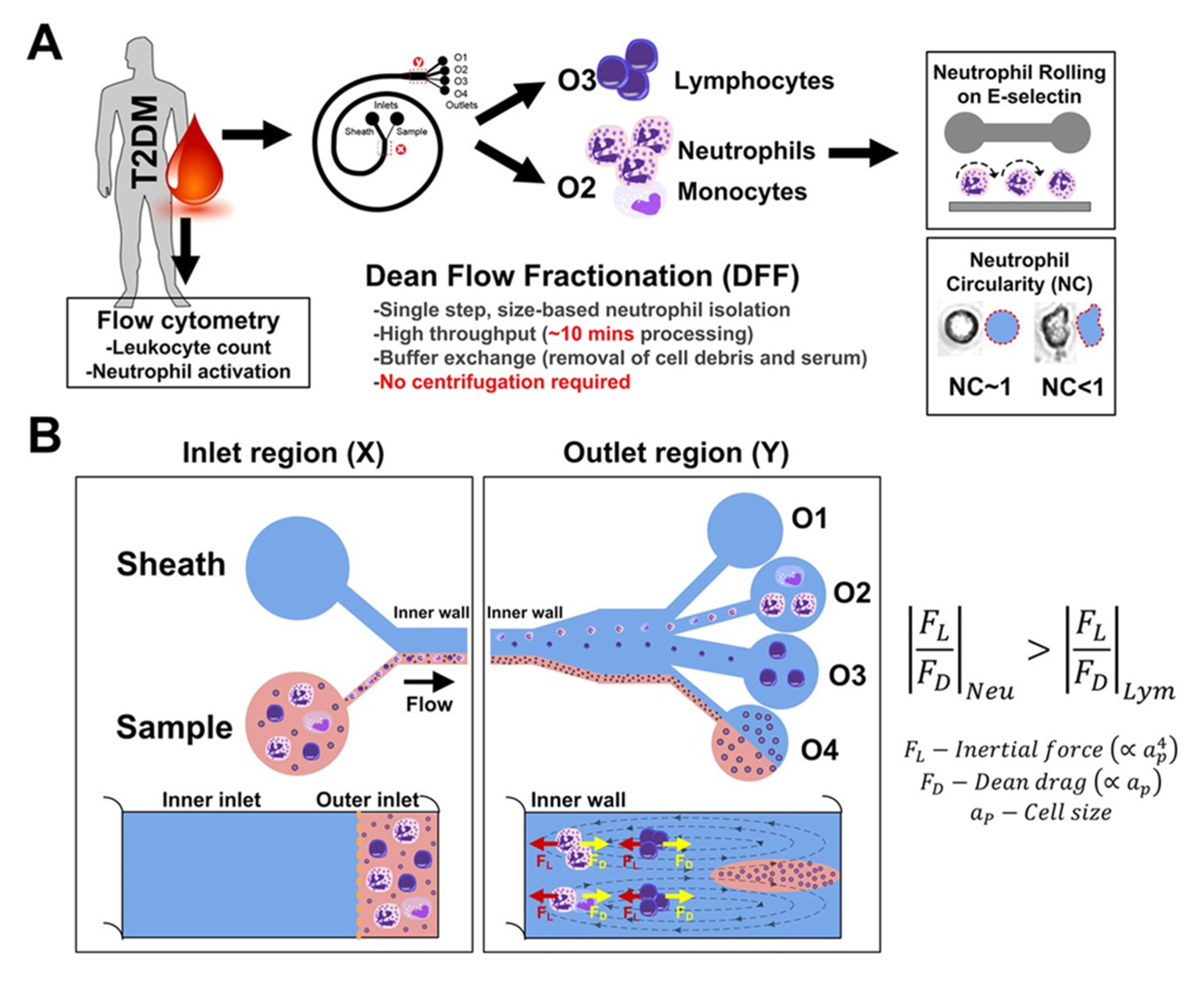
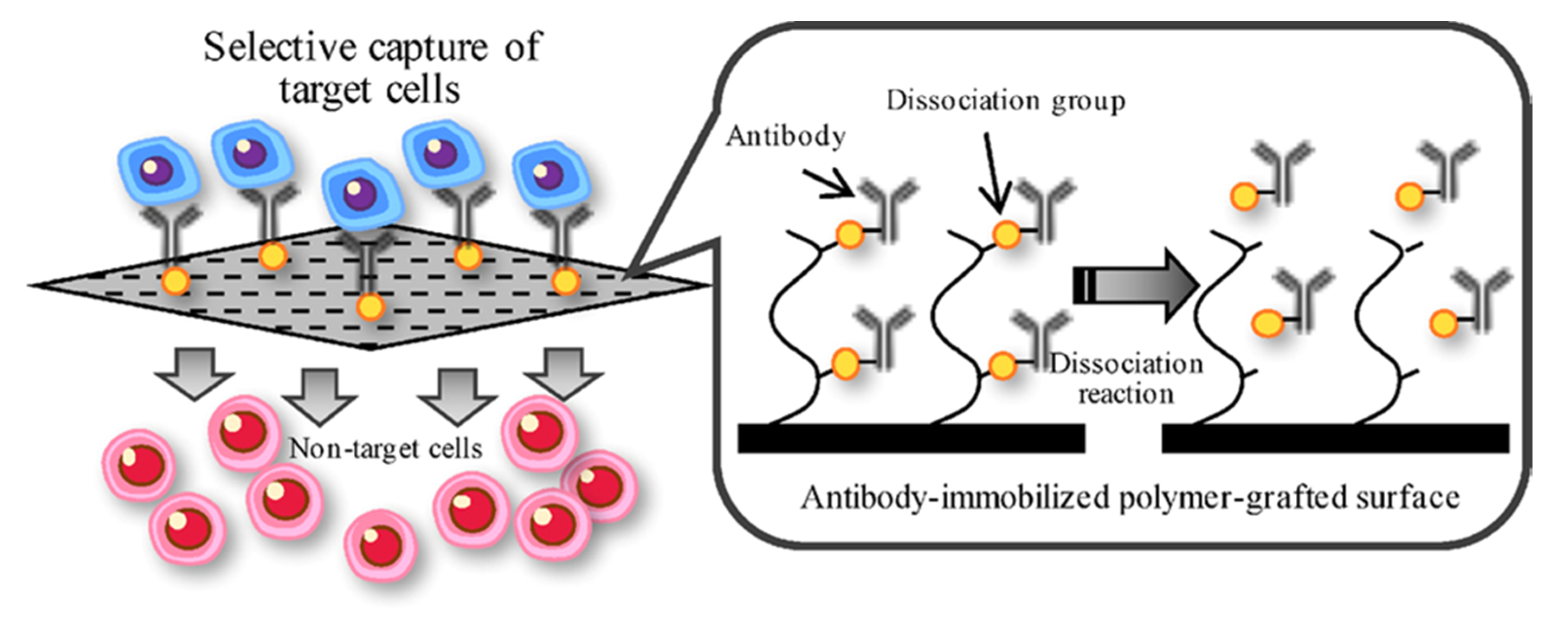

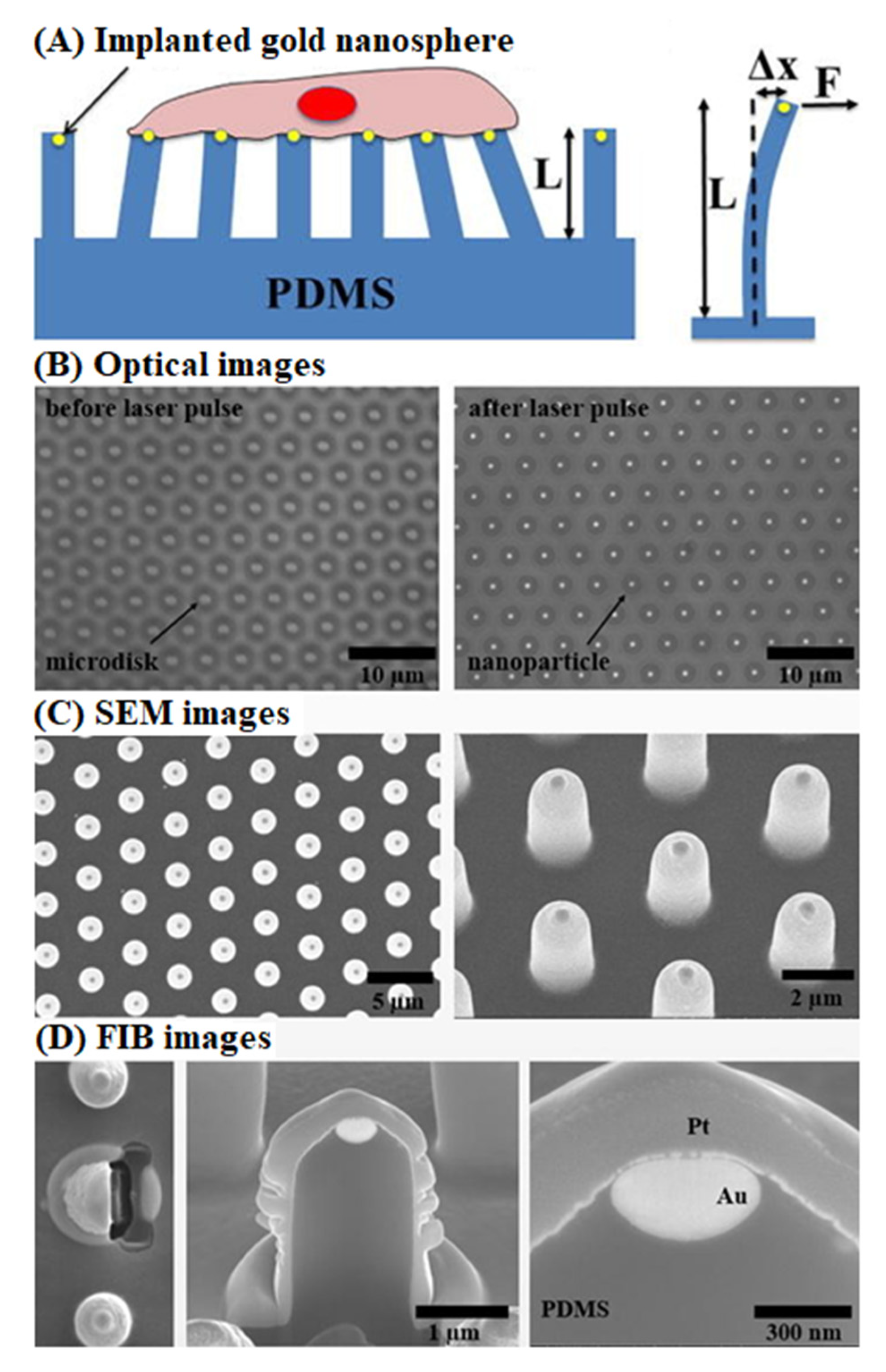
| Base Adhesion Layer | Described 1 Dimensions of Growth | Target Cells | Cellular Detection Method 2 | Described 1 Potential Applications | Ref. |
|---|---|---|---|---|---|
| Alginate | 3D | Mouse MSC | Fluorescent imaging | Cell encapsulation, pharmaceutical research, tissue engineering and regenerative medicine | [33] |
| Chondroitin sulfate | 3D | Porcine AC | DNA quantification and fluorescent imaging | Cartilage tissue engineering and regenerative medicine | [34] |
| Collagen | 3D | Human PCC | Fluorescence imaging | Disease modelling and drug screening | [35] |
| Cornstarch | 3D | Human osteosarcoma | Fluorescence imaging | Unspecified | [36] |
| Dextran | 3D | Rat BMSC and mouse EF | Fluorescence imaging | Cell encapsulation | [32] |
| Extracellular matrix | 3D | Human FH, human FSIC, human GS, human LDC, human SIC, and mouse SIC | Bright field and fluorescence imaging | Disease modelling, tissue regeneration and tissue repair | [37] |
| Elastin | 3D | Rat ASMC | Fluorescence imaging | Artificial vascular graft | [38] |
| Fibrinogen | 3D | Human cardiomyocytes | Fluorescence imaging and scanning electron microscopy | Tissue engineering and tissue regeneration | [39] |
| Fibronectin | 3D | Human BOSC, human CEC, and human FF | Fluorescence and phase contrast imaging | Wound dressing | [40] |
| Gelatin | 3D | Mouse SCTF | Fluorescence imaging | Drug delivery and tissue engineering | [41] |
| Hyaluronic acid | 3D | Mouse EF | Fluorescence imaging | Regenerative medicine and tissue engineering | [19] |
| Laminin | 2D | Human PSC-derived neurons | Fluorescence imaging | Regenerative medicine | [42] |
| Matrigel | 3D | Human PC | Fluorescence and phase contrast imaging | Disease modelling | [43] |
| Oxygen plasma 3 | 2D | Rat ADSC | Fluorescence imaging | Tissue engineering | [44] |
| Poly-D-lysine | 2D | Human ESC | Fluorescence imaging | Disease modelling | [45] |
| Poly-L-lysine | 3D | Rabbit marrow-derived MSC | Bright field imaging | Tissue engineering and tissue regeneration | [46] |
| Poly(ethylene glycol) | 3D | Human BGC, human DF, rat AGPC, and rat PIC | Fluorescence imaging | Cell delivery and tissue engineering | [20] |
| Poly(2-hydroxyethyl methacrylate) | 3D | Human MSC | Fluorescence imaging | Drug delivery and tissue engineering | [47] |
| Poly(N-isopropylacrylamide) | 2D | Mouse myoblast | Bright field and fluorescence imaging | Electronics for cell culture | [48] |
| Poly(vinyl alcohol) | 3D | Human BOSC | Fluorescence imaging | Tissue engineering | [49] |
| Recognition Element Type | Target Cell(s) | Described 1 Potential Applications(s) | Coupling Method/Linker 2 | Substrate | Cellular Detection Method 3 | Ref. |
|---|---|---|---|---|---|---|
| Amine plasma polymer | Bovine EC, human keratinocyte, human SF, mouse myoblasts and rat VSMC | Regenerative medicine | N/A | N/A | Fluorescence imaging | [82] |
| Antibody | EPC 4 | Biomedical devices | S-(11-Trichlorosilylundecanyl)-benzenethiosulfonate | 316L stainless steel | N/A | [83] |
| Antibody | Human EC and human VSMC | Implantable materials | Polyethylenimie + heparin + chitosan | Poly(ethylene terephalate) | Fluorescence imaging | [84] |
| Antibody | Mouse LC | Medical diagnostic and prognostic | Polyethylene glycol + poly(amidoamine) dendrimers | Epoxy-functionalized glass | Fluorescence imaging | [85] |
| Antibody | Mouse BM and mouse spleen | Cell therapy, immune therapy and regenerative medicine | Biotin + avidin + desthiobiotin | Polyethylene film | Fluorescence imaging | [86] |
| Antibody | Mouse BM | Cell therapy | Single stranded DNA + single stranded DNA | Polyethylene film | Fluorescence imaging | [87] |
| Antibody | Human EK | Medical diagnostics and implantable materials | Single stranded DNA + single stranded DNA + streptavidin + biotin | Cyclic olefin polymer film | Fluorescence imaging | [88] |
| Antibody | Human EK | Medical diagnostics and implantable materials | Single stranded DNA + single stranded DNA + streptavidin + biotin | Polycarbonate film | Fluorescence imaging | [88] |
| Antibody | Human BCC, human CCC, human HCC, human NSCLCC and human PCC | Medical diagnostics and monitoring | Thiol + DNA + biotin + avidin + biotin | Gold-plated PDMS | Fluorescence imaging | [89] |
| DNA (aptamer) | Human SMC, human UVEC and rat MSC-derived EPC | Implantable materials | Plasma polymerized allylamine | 316L stainless steel | Fluorescence imaging and QCM-D | [90] |
| Protein (E-selectin) | Human BA, human BC, human CA and human LA | Medical diagnostics | Sodium dodecanoate | Halloysite nanotubes | Fluorescence and non-fluorescence imaging | [91] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, J.V.L.; Ghafar-Zadeh, E. Biointerface Materials for Cellular Adhesion: Recent Progress and Future Prospects. Actuators 2020, 9, 137. https://doi.org/10.3390/act9040137
Nguyen JVL, Ghafar-Zadeh E. Biointerface Materials for Cellular Adhesion: Recent Progress and Future Prospects. Actuators. 2020; 9(4):137. https://doi.org/10.3390/act9040137
Chicago/Turabian StyleNguyen, John V. L., and Ebrahim Ghafar-Zadeh. 2020. "Biointerface Materials for Cellular Adhesion: Recent Progress and Future Prospects" Actuators 9, no. 4: 137. https://doi.org/10.3390/act9040137
APA StyleNguyen, J. V. L., & Ghafar-Zadeh, E. (2020). Biointerface Materials for Cellular Adhesion: Recent Progress and Future Prospects. Actuators, 9(4), 137. https://doi.org/10.3390/act9040137






