The Bovine Antimicrobial Peptide Lactoferricin Interacts with Polysialic Acid without Loss of Its Antimicrobial Activity against Escherichia coli
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Fractionation and Analysis of Neu5Ac Polymers
2.3. Electrophorese on Native Agarose Gel
2.4. ELISA
2.5. Bacterial Growth Assay
2.6. Statistical Analysis
3. Results and Discussion
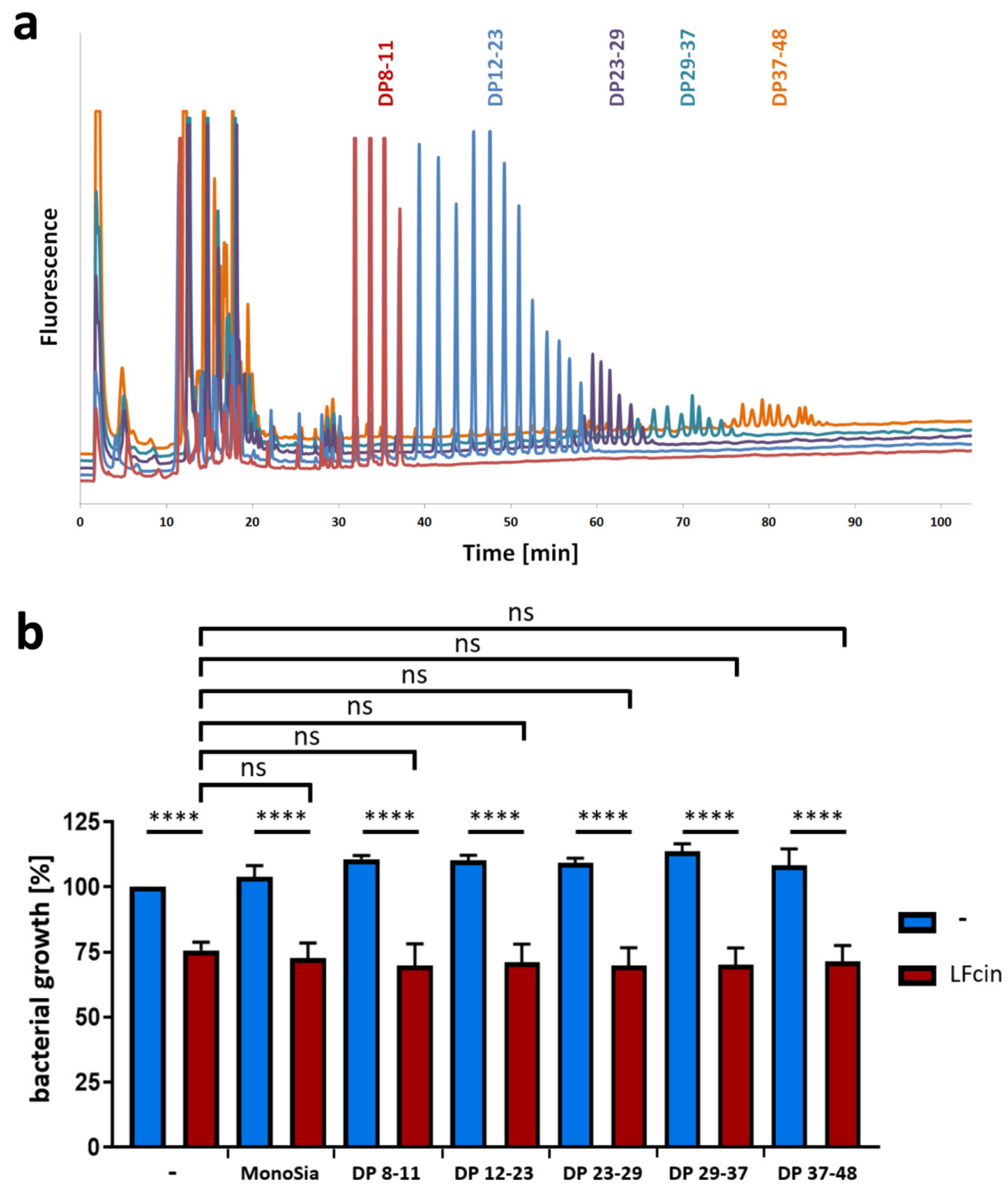
3.1. A Lower DP of PolySia Is Sufficient to Mediate the Binding to LFcin in Comparison to Lactoferrin
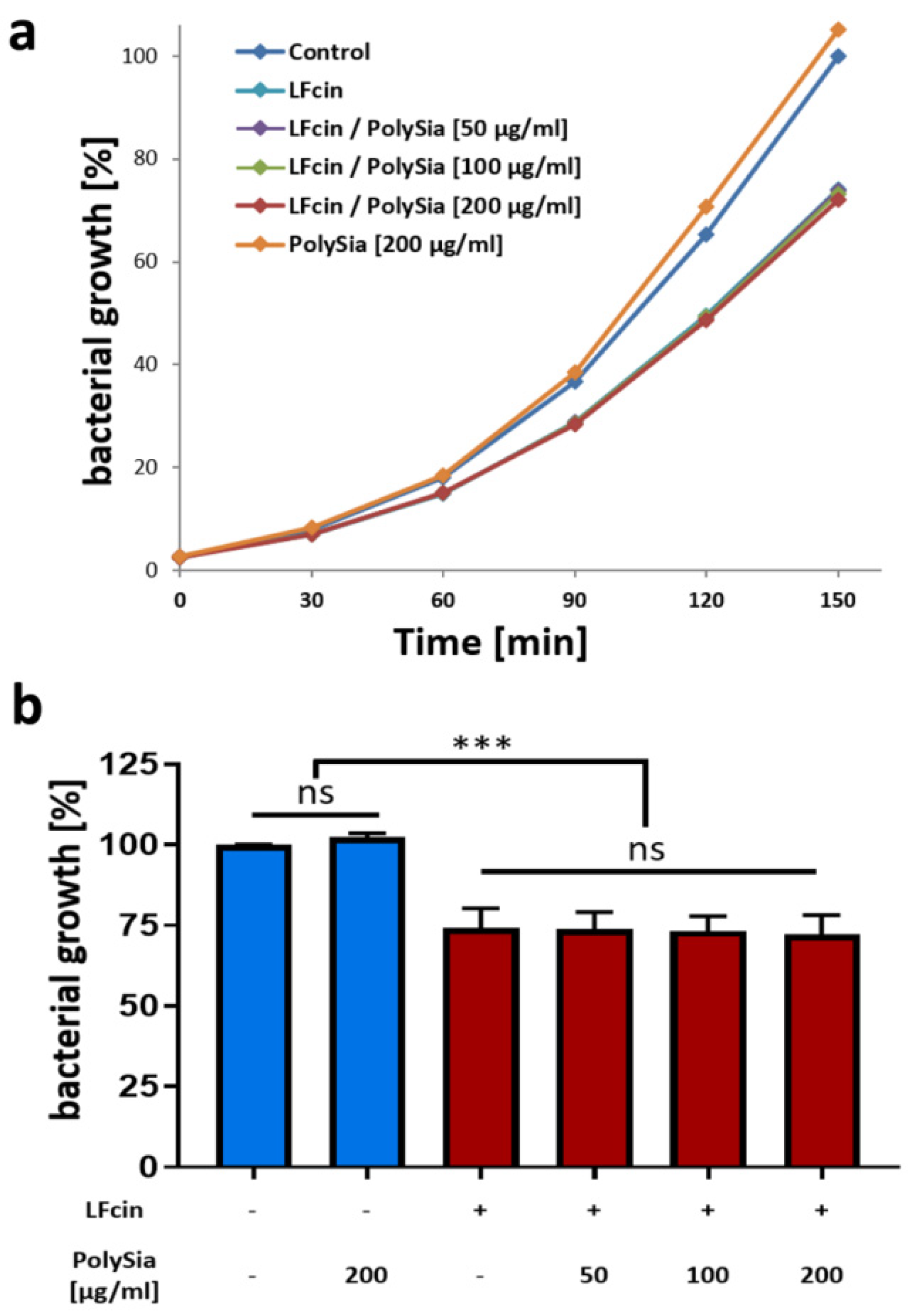
3.2. PolySia Has No Impact on the Antimicrobial Activity of LFcin
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, B.; Timilsena, Y.P.; Blanch, E.; Adhikari, B. Lactoferrin: Structure, function, denaturation and digestion. Crit. Rev. Food Sci. Nutr. 2017, 59, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kruzel, M.L.; Zimecki, M.; Actor, J.K. Lactoferrin in a context of inflammation-induced pathology. Front. Immunol. 2017, 8, 1438. [Google Scholar] [CrossRef] [PubMed]
- Okubo, K.; Kamiya, M.; Urano, Y.; Nishi, H.; Herter, J.M.; Mayadas, T.; Hirohama, D.; Suzuki, K.; Kawakami, H.; Tanaka, M.; et al. Lactoferrin suppresses neutrophil extracellular traps release in inflammation. EBioMedicine 2016, 10, 204–215. [Google Scholar] [CrossRef] [Green Version]
- Ammons, M.C.; Copie, V. Mini-review: Lactoferrin: A bioinspired, anti-biofilm therapeutic. Biofouling 2013, 29, 443–455. [Google Scholar] [CrossRef] [Green Version]
- Vogel, H.J. Lactoferrin, a bird’s eye view. Biochem. Cell Biol. 2012, 90, 233–244. [Google Scholar] [CrossRef]
- Gifford, J.L.; Hunter, H.N.; Vogel, H.J. Lactoferricin: A lactoferrin-derived peptide with antimicrobial, antiviral, antitumor and immunological properties. Cell. Mol. Life Sci. 2005, 62, 2588–2598. [Google Scholar] [CrossRef]
- Legrand, D. Overview of lactoferrin as a natural immune modulator. J. Pediatr-Us 2016, 173, S10–S15. [Google Scholar] [CrossRef] [Green Version]
- Levay, P.F.; Viljoen, M. Lactoferrin—A general-review. Haematologica 1995, 80, 252–267. [Google Scholar]
- Inoue, M.; Yamada, J.; Kitamura, N.; Shimazaki, K.; Andren, A.; Yamashita, T. Immunohistochemical localization of lactoferrin in bovine exocrine glands. Tissue Cell 1993, 25, 791–797. [Google Scholar] [CrossRef]
- Britigan, B.E.; Serody, J.S.; Cohen, M.S. The role of lactoferrin as an anti-inflammatory molecule. In Lactoferrin: Structure and Function; Hutchens, T.W., Rumball, S.V., Lönnerdal, B., Eds.; Springer: Boston, MA, USA, 1994; pp. 143–156. [Google Scholar]
- Kuhnle, A.; Lutteke, T.; Bornhofft, K.F.; Galuska, S.P. Polysialic acid modulates the binding of external lactoferrin in neutrophil extracellular traps. Biology 2019, 8, 20. [Google Scholar] [CrossRef] [Green Version]
- Kuhnle, A.; Veelken, R.; Galuska, C.E.; Saftenberger, M.; Verleih, M.; Schuppe, H.C.; Rudloff, S.; Kunz, C.; Galuska, S.P. Polysialic acid interacts with lactoferrin and supports its activity to inhibit the release of neutrophil extracellular traps. Carbohydr. Polym. 2019, 208, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Schnaar, R.L.; Gerardy-Schahn, R.; Hildebrandt, H. Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014, 94, 461–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, C. Chain length diversity of sialic acids and its biological significance. TIGG 2004, 16, 331–344. [Google Scholar] [CrossRef]
- Colley, K.J.; Kitajima, K.; Sato, C. Polysialic acid: Biosynthesis, novel functions and applications. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 498–532. [Google Scholar] [CrossRef]
- Sato, C.; Kitajima, K. Disialic, oligosialic and polysialic acids: Distribution, functions and related disease. J. Biochem. 2013, 154, 115–136. [Google Scholar] [CrossRef] [Green Version]
- Galuska, C.E.; Lütteke, T.; Galuska, S.P. Is polysialylated ncam not only a regulator during brain development but also during the formation of other organs? Biology 2017, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Werneburg, S.; Buettner, F.F.; Erben, L.; Mathews, M.; Neumann, H.; Muhlenhoff, M.; Hildebrandt, H. Polysialylation and lipopolysaccharide-induced shedding of e-selectin ligand-1 and neuropilin-2 by microglia and thp-1 macrophages. Glia 2016, 64, 1314–1330. [Google Scholar] [CrossRef]
- Kiermaier, E.; Moussion, C.; Veldkamp, C.T.; Gerardy-Schahn, R.; de Vries, I.; Williams, L.G.; Chaffee, G.R.; Phillips, A.J.; Freiberger, F.; Imre, R.; et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 2016, 351, 186–190. [Google Scholar] [CrossRef] [Green Version]
- Abe, C.; Yi, Y.; Hane, M.; Kitajima, K.; Sato, C. Acute stress-induced change in polysialic acid levels mediated by sialidase in mouse brain. Sci. Rep. 2019, 9, 9950. [Google Scholar] [CrossRef]
- Ulm, C.; Saffarzadeh, M.; Mahavadi, P.; Müller, S.; Prem, G.; Saboor, F.; Simon, P.; Middendorff, R.; Geyer, H.; Henneke, I.; et al. Soluble polysialylated ncam: A novel player of the innate immune system in the lung. Cell. Mol. Life Sci. 2013, 70, 3695–3708. [Google Scholar] [CrossRef]
- Zlatina, K.; Saftenberger, M.; Kuhnle, A.; Galuska, C.E.; Gartner, U.; Rebl, A.; Oster, M.; Vernunft, A.; Galuska, S.P. Polysialic acid in human plasma can compensate the cytotoxicity of histones. Int. J. Mol. Sci. 2018, 19, 1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yabe, U.; Sato, C.; Matsuda, T.; Kitajima, K. Polysialic acid in human milk. Cd36 is a new member of mammalian polysialic acid-containing glycoprotein. J. Biol. Chem. 2003, 278, 13875–13880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gluer, S.; Wunder, M.A.; Schelp, C.; Radtke, E.; Gerardy-Schahn, R. Polysialylated neural cell adhesion molecule serum levels in normal children. Pediatr. Res. 1998, 44, 915–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, P.; Bäumner, S.; Busch, O.; Röhrich, R.; Kaese, M.; Richterich, P.; Wehrend, A.; Müller, K.; Gerardy-Schahn, R.; Mühlenhoff, M.; et al. Polysialic acid is present in mammalian semen as a post-translational modification of the neural cell adhesion molecule ncam and the polysialyltransferase st8siaii. J. Biol. Chem. 2013, 288, 18825–18833. [Google Scholar] [CrossRef] [Green Version]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Bornhofft, K.F.; Galuska, S.P. Glycans as modulators for the formation and functional properties of neutrophil extracellular traps: Used by the forces of good and evil. Front. Immunol. 2019, 10, 959. [Google Scholar] [CrossRef] [Green Version]
- van Berkel, P.H.; Geerts, M.E.; van Veen, H.A.; Mericskay, M.; de Boer, H.A.; Nuijens, J.H. N-terminal stretch arg2, arg3, arg4 and arg5 of human lactoferrin is essential for binding to heparin, bacterial lipopolysaccharide, human lysozyme and DNA. Biochem. J. 1997, 328, 145–151. [Google Scholar] [CrossRef] [Green Version]
- Bellamy, W.; Takase, M.; Yamauchi, K.; Wakabayashi, H.; Kawase, K.; Tomita, M. Identification of the bactericidal domain of lactoferrin. Biochim. Biophys. Acta 1992, 1121, 130–136. [Google Scholar] [CrossRef]
- Kuwata, H.; Yip, T.T.; Tomita, M.; Hutchens, T.W. Direct evidence of the generation in human stomach of an antimicrobial peptide domain (lactoferricin) from ingested lactoferrin. BBA-Protein Struct. Mol. 1998, 1429, 129–141. [Google Scholar] [CrossRef]
- Britigan, B.E.; Hayek, M.B.; Doebbeling, B.N.; Fick, R.B., Jr. Transferrin and lactoferrin undergo proteolytic cleavage in the pseudomonas aeruginosa-infected lungs of patients with cystic fibrosis. Infect. Immun. 1993, 61, 5049–5055. [Google Scholar]
- Yamauchi, K.; Tomita, M.; Giehl, T.J.; Ellison, R.T., 3rd. Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect. Immun. 1993, 61, 719–728. [Google Scholar] [PubMed]
- Bellamy, W.; Takase, M.; Wakabayashi, H.; Kawase, K.; Tomita, M. Antibacterial spectrum of lactoferricin b, a potent bactericidal peptide derived from the n-terminal region of bovine lactoferrin. J. Appl. Bacteriol. 1992, 73, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Bruni, N.; Capucchio, M.T.; Biasibetti, E.; Pessione, E.; Cirrincione, S.; Giraudo, L.; Corona, A.; Dosio, F. Antimicrobial activity of lactoferrin-related peptides and applications in human and veterinary medicine. Molecules 2016, 21, 752. [Google Scholar] [CrossRef] [PubMed]
- Saffarzadeh, M.; Juenemann, C.; Queisser, M.A.; Lochnit, G.; Barreto, G.; Galuska, S.P.; Lohmeyer, J.; Preissner, K.T. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: A predominant role of histones. PLoS ONE 2012, 7, e32366. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; von der Ohe, M.; Schulze, C.; Bian, S.; Makhina, T.; Loers, G.; Kleene, R.; Schachner, M. Functional role of the interaction between polysialic acid and extracellular histone h1. J. Neurosci. 2010, 30, 12400–12413. [Google Scholar] [CrossRef]
- Shabir, U.; Ali, S.; Magray, A.R.; Ganai, B.A.; Firdous, P.; Hassan, T.; Nazir, R. Fish antimicrobial peptides (amp’s) as essential and promising molecular therapeutic agents: A review. Microb. Pathog. 2018, 114, 50–56. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Bhat, R.A.H.; Pande, A. Antimicrobial peptides of fish: Innocuous alternatives to antibiotics. Rev. Aquac. 2018. [Google Scholar] [CrossRef]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [Green Version]
- Parseghian, M.H.; Luhrs, K.A. Beyond the walls of the nucleus: The role of histones in cellular signaling and innate immunity. Biochem. Cell Biol. 2006, 84, 589–604. [Google Scholar] [CrossRef]
- Park, I.Y.; Park, C.B.; Kim, M.S.; Kim, S.C. Parasin i, an antimicrobial peptide derived from histone h2a in the catfish, parasilurus asotus. FEBS Lett. 1998, 437, 258–262. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, H.; Koyama, T.; Conlon, J.M.; Yamakura, F.; Iwamuro, S. Antimicrobial action of histone h2b in escherichia coli: Evidence for membrane translocation and DNA-binding of a histone h2b fragment after proteolytic cleavage by outer membrane proteinase t. Biochimie 2008, 90, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Silphaduang, U.; Hincke, M.T.; Nys, Y.; Mine, Y. Antimicrobial proteins in chicken reproductive system. Biochem. Biophys. Res. Commun. 2006, 340, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Li, G.H.; Mine, Y.; Hincke, M.T.; Nys, Y. Isolation and characterization of antimicrobial proteins and peptide from chicken liver. J. Pept. Sci. 2007, 13, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Hiemstra, P.S.; Eisenhauer, P.B.; Harwig, S.S.; van den Barselaar, M.T.; van Furth, R.; Lehrer, R.I. Antimicrobial proteins of murine macrophages. Infect. Immun. 1993, 61, 3038–3046. [Google Scholar]
- Galuska, S.P.; Galuska, C.E.; Tharmalingam, T.; Zlatina, K.; Prem, G.; Husejnov, F.C.O.; Rudd, P.M.; Vann, W.F.; Reid, C.; Vionnet, J.; et al. In vitro generation of polysialylated cervical mucins by bacterial polysialyltransferases to counteract cytotoxicity of extracellular histones. FEBS J. 2017, 284, 1688–1699. [Google Scholar] [CrossRef] [Green Version]
- Galuska, C.E.; Dambon, J.A.; Kuhnle, A.; Bornhofft, K.F.; Prem, G.; Zlatina, K.; Lutteke, T.; Galuska, S.P. Artificial polysialic acid chains as sialidase-resistant molecular-anchors to accumulate particles on neutrophil extracellular traps. Front. Immunol. 2017, 8, 1229. [Google Scholar] [CrossRef] [Green Version]
- Zlatina, K.; Lütteke, T.; Galuska, S.P. Individual impact of distinct polysialic acid chain lengths on the cytotoxicity of histone h1, h2a, h2b, h3 and h4. Polymers 2017, 9, 720. [Google Scholar] [CrossRef] [Green Version]
- Zlatina, K.; Galuska, S.P. Polysialic acid modulates only the antimicrobial properties of distinct histones. ACS Omega 2019, 4, 1601–1610. [Google Scholar] [CrossRef]
- Hara, S.; Yamaguchi, M.; Takemori, Y.; Furuhata, K.; Ogura, H.; Nakamura, M. Determination of mono-o-acetylated n-acetylneuraminic acids in human and rat sera by fluorometric high-performance liquid chromatography. Anal. Biochem. 1989, 179, 162–166. [Google Scholar] [CrossRef]
- Hara, S.; Takemori, Y.; Yamaguchi, M.; Nakamura, M.; Ohkura, Y. Fluorometric high-performance liquid chromatography of n-acetyl- and n-glycolylneuraminic acids and its application to their microdetermination in human and animal sera, glycoproteins, and glycolipids. Anal. Biochem. 1987, 164, 138–145. [Google Scholar] [CrossRef]
- Inoue, S.; Lin, S.L.; Lee, Y.C.; Inoue, Y. An ultrasensitive chemical method for polysialic acid analysis. Glycobiology 2001, 11, 759–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, R.; Yokota, H.; Kim, S.H. Electrophoresis of proteins and protein-protein complexes in a native agarose gel. Anal. Biochem. 2000, 282, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, P.M.; Zhou, N.; Shan, X.; Arrowsmith, C.H.; Vogel, H.J. Three-dimensional solution structure of lactoferricin b, an antimicrobial peptide derived from bovine lactoferrin. Biochemistry 1998, 37, 4288–4298. [Google Scholar] [CrossRef]
- Hallenbeck, P.C.; Vimr, E.R.; Yu, F.; Bassler, B.; Troy, F.A. Purification and properties of a bacteriophage-induced endo-n-acetylneuraminidase specific for poly-alpha-2,8-sialosyl carbohydrate units. J. Biol. Chem. 1987, 262, 3553–3561. [Google Scholar]
- Rutishauser, U.; Watanabe, M.; Silver, J.; Troy, F.A.; Vimr, E.R. Specific alteration of ncam-mediated cell adhesion by an endoneuraminidase. J. Cell Biol. 1985, 101, 1842–1849. [Google Scholar] [CrossRef] [Green Version]
- Curreli, S.; Arany, Z.; Gerardy-Schahn, R.; Mann, D.; Stamatos, N.M. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-t lymphocyte interactions. J. Biol. Chem. 2007, 282, 30346–30356. [Google Scholar] [CrossRef] [Green Version]
- Haiwen, Z.; Rui, H.; Bingxi, Z.; Qingfeng, G.; Jifeng, Z.; Xuemei, W.; Beibei, W. Oral administration of bovine lactoferrin-derived lactoferricin (lfcin) B could attenuate enterohemorrhagic Escherichia coli O157:H7 induced intestinal disease through improving intestinal barrier function and microbiota. J. Agric. Food Chem. 2019, 67, 3932–3945. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kühnle, A.; Galuska, C.E.; Zlatina, K.; Galuska, S.P. The Bovine Antimicrobial Peptide Lactoferricin Interacts with Polysialic Acid without Loss of Its Antimicrobial Activity against Escherichia coli. Animals 2020, 10, 1. https://doi.org/10.3390/ani10010001
Kühnle A, Galuska CE, Zlatina K, Galuska SP. The Bovine Antimicrobial Peptide Lactoferricin Interacts with Polysialic Acid without Loss of Its Antimicrobial Activity against Escherichia coli. Animals. 2020; 10(1):1. https://doi.org/10.3390/ani10010001
Chicago/Turabian StyleKühnle, Andrea, Christina E. Galuska, Kristina Zlatina, and Sebastian P. Galuska. 2020. "The Bovine Antimicrobial Peptide Lactoferricin Interacts with Polysialic Acid without Loss of Its Antimicrobial Activity against Escherichia coli" Animals 10, no. 1: 1. https://doi.org/10.3390/ani10010001
APA StyleKühnle, A., Galuska, C. E., Zlatina, K., & Galuska, S. P. (2020). The Bovine Antimicrobial Peptide Lactoferricin Interacts with Polysialic Acid without Loss of Its Antimicrobial Activity against Escherichia coli. Animals, 10(1), 1. https://doi.org/10.3390/ani10010001




