Starch-Rich Diet Induced Rumen Acidosis and Hindgut Dysbiosis in Dairy Cows of Different Lactations
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Setup, Animals, and Feeding
2.2. Fecal Sampling and Fermentation Parameters
2.3. DNA Extraction and Sequencing
2.4. Bioinformatic Analysis
2.5. Statistical Analysis
3. Results
3.1. Effects of Feeding an Increased Starch Level on Fecal pH and SCFA
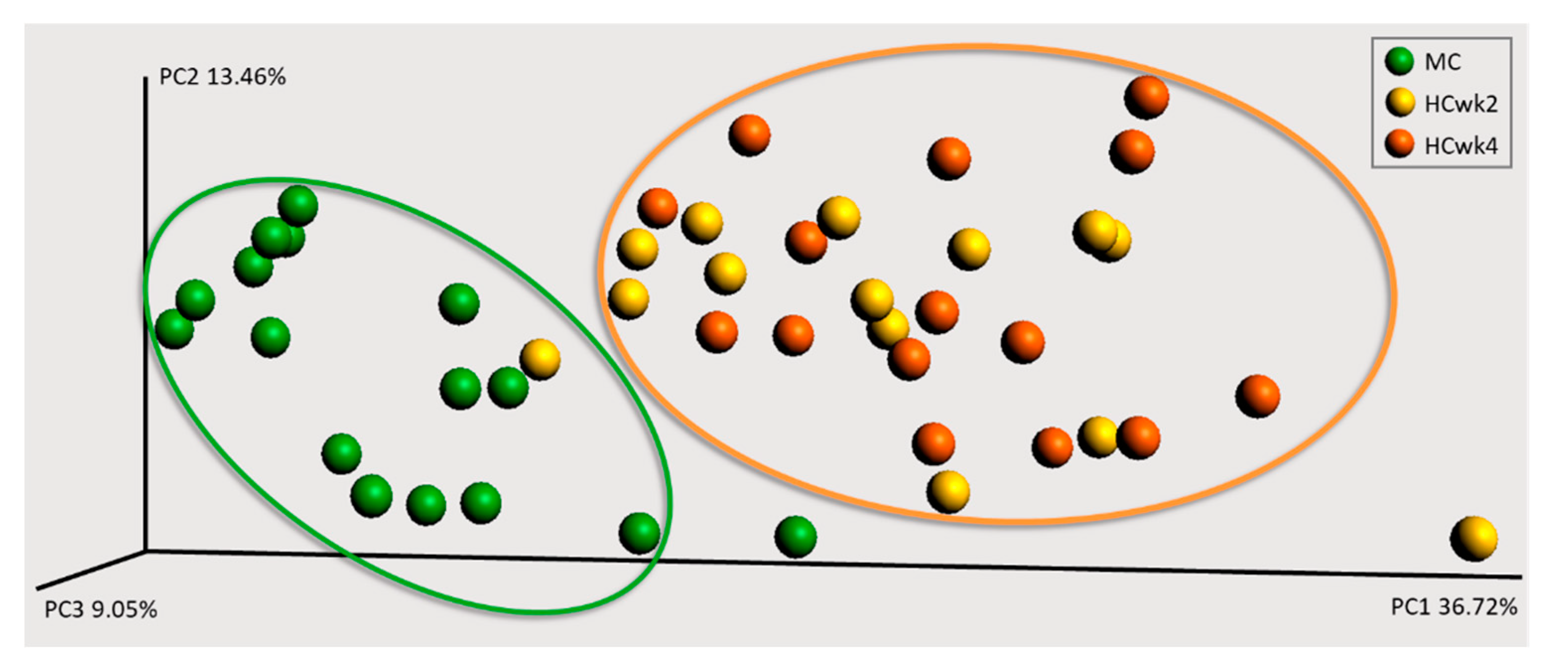
3.2. Effects of Feeding an Increased Starch Level on the Fecal Microbiome
3.3. Effects of Lactation Number on Performance, Ruminal pH, Fecal pH, and SCFA
3.4. Effects of Lactation Number on the Fecal Microbiome
4. Discussion
4.1. High-Concentrate Feeding Causes Hindgut Dysbiosis
4.2. Rumen Acidosis and Hindgut Dysbiosis in Different Lactations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meissner, S.; Hagen, F.; Deiner, C.; Günzel, D.; Greco, G.; Shen, Z.; Aschenbach, J.R. Key role of short-chain fatty acids in epithelial barrier failure during ruminal acidosis. J. Dairy Sci. 2017, 100, 6662–6675. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Khafipour, E.; Krause, D.O.; Kroeker, A.; Rodriguez-Lecompte, J.C.; Gozho, G.N.; Plaizier, J.C. Effects of subacute ruminal acidosis challenges on fermentation and endotoxins in the rumen and hindgut of dairy cows. J. Dairy Sci. 2012, 95, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.C.; Krause, D.O.; Gozho, G.N.; McBride, B.W. Subacute ruminal acidosis in dairy cows: The physiological causes, incidence and consequences. Vet. J. 2008, 176, 21–31. [Google Scholar] [CrossRef]
- Steele, M.A.; Croom, J.; Kahler, M.; AlZahal, O.; Hook, S.E.; Plaizier, K.; McBride, B.W. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R1515–R1523. [Google Scholar] [CrossRef] [Green Version]
- Gressley, T.F.; Hall, M.B.; Armentano, L.E. Ruminant nutrition symposium: Productivity, digestion, and health responses to hindgut acidosis in ruminants. J. Anim. Sci. 2011, 89, 1120–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmanuel, D.G.V.; Madsen, K.L.; Churchill, T.A.; Dunn, S.M.; Ametaj, B.N. Acidosis and Lipopolysaccharide from Escherichia coli B:055 cause hyperpermeability of rumen and colon tissues. J. Dairy Sci. 2007, 90, 5552–5557. [Google Scholar] [CrossRef] [Green Version]
- Khafipour, E.; Krause, D.O.; Plaizier, J.C. Alfalfa pellet-induced subacute ruminal acidosis in dairy cows increases bacterial endotoxin in the rumen without causing inflammation. J. Dairy Sci. 2009, 92, 1712–1724. [Google Scholar] [CrossRef] [Green Version]
- Mao, S.; Zhang, M.; Liu, J.; Zhu, W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015, 5, 16116. [Google Scholar] [CrossRef] [Green Version]
- Humer, E.; Kohl-Parisini, A.; Gruber, L.; Gasteiner, J.; Abdel-Raheem, S.M.; Zebeli, Q. Long-term reticuloruminal pH dynamics and markers of liver health in early-lactating cows of various parities fed diets differing in grain processing. J. Dairy Sci. 2015, 98, 6433–6448. [Google Scholar] [CrossRef]
- Stauder, A.; Humer, E.; Neubauer, V.; Reisinger, N.; Kaltenegger, A.; Zebeli, Q. Distinct responses in feed sorting, chewing behavior, and ruminal acidosis risk between primiparous and multiparous Simmental cows fed diets differing in forage and starch level. J. Dairy Sci. 2020, 103, 8469–8481. [Google Scholar] [CrossRef] [PubMed]
- Penner, G.B.; Steele, M.A.; Aschenbach, J.R.; McBride, B.W. Ruminant nutrition symposium: Molecular adaptation of ruminal epithelia to highly fermentable diets. J. Anim. Sci. 2011, 89, 1108–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neubauer, V.; Humer, E.; Kröger, I.; Meißl, A.; Reisinger, N.; Zebeli, Q. Technical note: Changes in rumen mucosa thickness measured by transabdominal ultrasound as a noninvasive method to diagnose subacute rumen acidosis in dairy cows. J. Dairy Sci. 2018, 101, 2650–2654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Lin, X.; Wang, Z.; Hou, Q.; Wang, Y.; Hu, Z. High-production dairy cattle exhibit different rumen and fecal bacterial community and rumen metabolite profile than low-production cattle. Microbiol. Open 2019, 8, 1–12. [Google Scholar] [CrossRef]
- Liu, C.; Meng, Q.; Chen, Y.; Xu, M.; Shen, M.; Gao, R.; Gan, S. Role of age-related shifts in rumen bacteria and methanogens in methane production in cattle. Front. Microbiol. 2017, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kröger, I.; Humer, E.; Neubauer, V.; Reisinger, N.; Zebeli, Q. Feeding diets moderate in physically effective fibre alters eating and feed sorting patterns without improving ruminal pH, but impaired liver health in dairy cows. Animals 2019, 9, 128. [Google Scholar] [CrossRef] [Green Version]
- Qumar, M.; Khiaosa-Ard, R.; Pourazad, P.; Wetzels, S.U.; Klevenhusen, F.; Kandler, W.; Aschenbach, J.R.; Zebeli, Q. Evidence of in vivo absorption of lactate and modulation of Short chain fatty acid absorption from the reticulorumen of non-lactating cattle fed high concentrate diets. PLoS ONE 2016, 11, e0164192. [Google Scholar] [CrossRef]
- Peterson, P.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.; Bonazzi, V.; McEwen, J.; Wetterstrand, K.; Deal, C.; et al. The NIH human microbiome project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef] [Green Version]
- EMBL-EBI ENA (European Nucleotide Archive). Available online: https://www.ebi.ac.uk/ena/browser/home (accessed on 1 August 2020).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- QIIME Tutorials. Available online: http://qiime.org/tutorials/ (accessed on 1 May 2019).
- Navas-Molina, J.A.; Peralta-Sánchez, J.M.; González, A.; McMurdie, P.J.; Vázquez-Baeza, Y.; Xu, Z.; Ursell, L.K.; Lauber, C.; Zhou, H.; Jin Song, S.; et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013, 531, 371–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemos, L.N.; Fulthorpe, R.R.; Triplett, E.W.; Roesch, L.F.W. Rethinking microbial diversity analysis in the high throughput sequencing era. J. Microbiol. Methods 2011, 86, 42–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Stamatis, D.; Bertsch, J.; Ovchinnikova, G.; Verezemska, O.; Isbandi, M.; Thomas, A.D.; Ali, R.; Sharma, K.; Kyrpides, N.C.; et al. Genomes OnLine Database (GOLD) v.6: Data updates and feature enhancements. Nucleic Acids Res. 2017, 45, D446–D456. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SILVA ribosomal RNA database, Max Planck Institute for Marine Microbiology and Jacobs University, Bremen, Germany. Available online: https://www.arb-silva.de (accessed on 1 May 2019).
- Akoglu, H. User’s guide to correlation coefficients. Turkish J. Emerg. Med. 2018, 18, 91–93. [Google Scholar] [CrossRef]
- Mao, S.; Zhang, R.; Wang, D.; Zhu, W. The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. BMC Vet. Res. 2012, 8, 237. [Google Scholar] [CrossRef] [Green Version]
- Sulzberger, S.A.; Kalebich, C.C.; Melnichenko, S.; Cardoso, F.C. Effects of clay after a grain challenge on milk composition and on ruminal, blood, and fecal pH in Holstein cows. J. Dairy Sci. 2016, 99, 8028–8040. [Google Scholar] [CrossRef] [Green Version]
- Neubauer, V.; Petri, R.; Humer, E.; Kröger, I.; Mann, E.; Reisinger, N.; Wagner, M.; Zebeli, Q. High-grain diets supplemented with phytogenic compounds or autolyzed yeast modulate ruminal bacterial community and fermentation in dry cows. J. Dairy Sci. 2018, 101, 2335–2349. [Google Scholar] [CrossRef]
- von Engelhardt, W.; Breves, G.; Diener, M.; Gäbel, G. Physiologie der Haustiere, 5th ed.; Enke Verlag: Stuttgart, Germany, 2015. [Google Scholar]
- Puniya, A.K.; Singh, R.; Kamra, D.N. Rumen Microbiology: From Evolution to Revolution, 1st ed.; Springer: New Delhi, India, 2015; ISBN 978-81-322-2400-6. [Google Scholar]
- Nobs, S.P.; Tuganbaev, T.; Elinav, E. Microbiome diurnal rhythmicity and its impact on host physiology and disease risk. EMBO Rep. 2019, 20, 1–15. [Google Scholar] [CrossRef]
- Noel, S.J.; Olijhoek, D.W.; McLean, F.; Løvendahl, P.; Lund, P.; Højberg, O. Rumen and fecal microbial community structure of holstein and Jersey dairy cows as affected by breed, diet, and residual feed intake. Animals 2019, 9, 498. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Han, Y.; Fan, H.; Zhao, J.; Wang, J. Characterization of the microbial communities along the gastrointestinal tract of sheep by 454 pyrosequencing analysis. Asian Australas. J. Anim. Sci. 2016, 30, 100–110. [Google Scholar] [CrossRef]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.S.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.L.; Tringe, S.G. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, J.B.; Wilson, D.B. Why are ruminal cellulolytic bacteria unable to digest cellulose at low pH? J. Dairy Sci. 1996, 79, 1503–1509. [Google Scholar] [CrossRef]
- Stanley, K.; Jones, K. Cattle and sheep farms as reservoirs of Campylobacter. J. Appl. Microbiol. 2003, 94, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, M. The Prokaryotes Vol 3: Archaea. Bacteria: Firmicutes, Actinomycetes, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; ISBN 978-0-387-25492-0. [Google Scholar]
- Zinicola, M.; Lima, F.; Lima, S.; Machado, V.; Gomez, M.; Döpfer, D.; Guard, C.; Bicalho, R. Altered microbiomes in bovine digital dermatitis lesions, and the gut as a pathogen reservoir. PLoS ONE 2015, 10, e0120504. [Google Scholar] [CrossRef]
- Neubauer, V.; Humer, E.; Kröger, I.; Braid, T.; Wagner, M.; Zebeli, Q. Differences between pH of indwelling sensors and the pH of fluid and solid phase in the rumen of dairy cows fed varying concentrate levels. J. Anim. Physiol. Anim. Nutr. 2018, 102, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Zebeli, Q.; Ghareeb, K.; Humer, E.; Metzler-Zebeli, B.U.; Besenfelder, U. Nutrition, rumen health and inflammation in the transition period and their role on overall health and fertility in dairy cows. Res. Vet. Sci. 2015, 103, 126–136. [Google Scholar] [CrossRef]
- Van Soest, P.J. Nutritional Ecology of the Ruminant, 2nd ed.; Cornell University Press: Ithaca, NY, USA, 1994. [Google Scholar]
- Duarte, A.; McCullough, D.R.; Weckerly, F.W. Does rumen-reticulum capacity correlate with body size or age in black-tailed deer? Eur. J. Wildl. Res. 2011, 57, 1131–1136. [Google Scholar] [CrossRef]
- Aschenbach, J.R.; Penner, G.B.; Stumpff, F.; Gäbel, G. Ruminant nutrition symposium: Role of fermentation acid absorption in the regulation of ruminal pH. J. Anim. Sci. 2011, 89, 1092–1107. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Wang, Y.; Luo, H.; Qiu, W.; Zhang, H.; Hu, L.; Wang, Y.; Dong, G.; Guo, G. The association between inflammaging and age-related changes in the ruminal and fecal microbiota among lactating holstein cows. Front. Microbiol. 2019, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]



| Parameter | Feeding Phase 1 | SEM | p-Value Phase | |||
|---|---|---|---|---|---|---|
| MC | HCwk2 | HCwk3 | HCwk4 | |||
| Fecal pH (mean) | 7.78 a | 7.51 b | 7.45 b,c | 7.39 c | 0.038 | <0.001 |
| Total SCFA (mmol/L) | 55.7 b | 88.9 a | 83.0 a | 86.0 a | 6.07 | <0.001 |
| Acetate (%) | 75.5 a | 73.9 b | 74.8 | 74.4 | 0.77 | 0.08 |
| Propionate (%) | 14.3 a | 13.4 b | 13.8 a | 13.1 b | 0.29 | 0.01 |
| Butyrate (%) | 4.3 c | 5.3a | 5.2 a | 4.8 b | 0.17 | <0.001 |
| Valerate (%) | 1.14 a | 1.11 a,b | 1.01 b | 0.99 b | 0.05 | 0.05 |
| Caproate (%) | 0.16 b | 2.09 | 0.46 b | 2.49 a | 0.819 | 0.05 |
| iso-Butyrate (%) | 3.9 | 3.9 | 4.3 | 3.8 | 0.24 | 0.48 |
| iso-Valerate (%) | 0.84 a | 0.36 | 0.46 b | 0.38 b | 0.047 | <0.001 |
| Parameter | Feeding Phase 1 | SEM | p-Value Phase | ||
|---|---|---|---|---|---|
| MC | HCwk2 | HCwk4 | |||
| Goods coverage | 0.920 b | 0.931 a | 0.934 a | 0.0026 | <0.001 |
| Shannon | 8.85 a | 8.24 b | 8.19 b | 0.092 | <0.001 |
| Simpson | 0.992 a | 0.983 b | 0.984 b | 0.0017 | <0.001 |
| Chao1 | 2813 a | 2418 b | 2349 b | 83.2 | <0.001 |
| Singletons | 798 a | 687 b | 656 b | 26.6 | <0.001 |
| Parameter | MC | HCwk1 | HCwk2 | HCwk3 | HCwk4 | SEM | p-Value Lact. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2ndL | 3rdL | ≥4 L | 2ndL | 3rdL | ≥4 L | 2ndL | 3rdL | ≥4 L | 2ndL | 3rdL | ≥4 L | 2ndL | 3rdL | ≥4 L | |||
| Performance and Chewing | |||||||||||||||||
| Milk yield (kg/day) | 29.9 (b) | 34.4 (a) | 34.5 (a) | 30.6 b | 35.3 a | 35.2 (a) | 31.7 (b) | 36.2 (a) | 36.6 (a) | 31.4 b | 36.9 a | 36.2 a | 31.0 b | 34.9 (a) | 36.6 a | 1.66 | <0.001 |
| DMI (kg/day) | 20.7 b | 23.5 a | 22.1 | 21.3 b | 24.3 a | 24.8 a | 21.9 (b) | 24.4 | 25.0 (a) | 22.9 b | 25.6 a | 24.9 | 22.8 b | 26.0 a | 25.6 a | 0.98 | <0.001 |
| DMI/BW (kg/kg) | 3.2 a | 3.1 a | 2.7 b | 3.3 | 3.2 | 3.1 | 3.4 (a) | 3.2 | 3.1 (b) | 3.5 a | 3.4 (a) | 3.1 b | 3.5 a | 3.5 a | 3.2 b | 0.15 | <0.001 |
| Milk yield/DMI (kg/kg) | 1.5 | 1.5 | 1.6 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.4 | 1.5 | 1.5 | 1.4 | 1.3 | 1.5 | 0.09 | n.s. |
| Rumination min/day | 569 | 581 | 573 | 522 | 530 | 556 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | 533 | 548 | 553 | 20.7 | n.s. |
| Eating min/day | 377 a | 399 a | 292 b | 333 | 353 (a) | 288 (b) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | 332 | 332 | 298 | 38.7 | 0.05 |
| Rumination min/kg DMI | 27.9 (a) | 25.2 (b) | 28.1 (a) | 25.6 a | 22.5 b | 23.3 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | 23.1 | 20.4 | 21.0 | 1.07 | 0.04 |
| Eating min/kg DMI | 18.4 a | 16.9 | 14.2b | 16.8 a | 14.9 | 11.8 b | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | 14.3 | 12.2 | 11.3 | 1.68 | 0.02 |
| Rumen | |||||||||||||||||
| Mean pH | 6.39 | 6.30 | 6.70 | 6.27 | 6.18 | 6.56 | 6.26 | 6.20 | 6.50 | 6.22 | 6.17 | 6.46 | 6.24 | 6.14 | 6.42 | 0.220 | <0.001 1 |
| Minimum pH | 6.00 | 5.94 | 6.33 | 5.78 | 5.72 | 6.08 | 5.74 | 5.68 | 6.03 | 5.72 | 5.61 | 6.02 | 5.82 | 5.65 | 5.98 | 0.211 | <0.001 1 |
| Maximum pH | 6.71 b | 6.49 b | 7.09 a | 6.75 b | 6.53 b | 7.05 a | 6.70 | 6.49 b | 6.98 a | 6.67 | 6.50 b | 6.94 a | 6.57 | 6.42 b | 6.87 a | 0.110 | <0.001 |
| minutes pH<6.0 | 204 | 454 a | 21 b | 380 | 640 a | 151 b | 370 | 598 a | 99 b | 438 | 636 a | 127 b | 480 | 703 a | 96 b | 137.7 | <0.001 |
| Milk content | |||||||||||||||||
| Fat (%) | 3.8 | 3.9 | 3.9 | n.a. | n.a. | n.a. | 3.9 | 3.6 | 3.5 | 5.1 (a) | 4.0 (b) | 3.9 (b) | 3.6 | 3.7 | 3.4 | 0.44 | 0.08 |
| Protein (%) | 3.4 | 3.4 | 3.2 | n.a. | n.a. | n.a. | 3.5 | 3.5 | 3.5 | 3.5 | 3.6 | 3.5 | 3.6 | 3.6 | 3.6 | 0.10 | n.s. |
| Lactose (%) | 4.9 | 4.8 | 4.8 | n.a. | n.a. | n.a. | 4.8 | 4.7 | 4.7 | 4.8 | 4.7 | 4.7 | 4.8 | 4.7 | 4.7 | 0.06 | 0.06 |
| Milk urea nitrogen (mg/dL) | 26.9 a | 26.4 a | 20.5 b | n.a. | n.a. | n.a. | 19.0 | 18.2 | 16.6 | 21.9 | 21.5 | 18.3 | 25.5 | 22.2 | 22.5 | 1.53 | 0.01 |
| Urea/DMI (mg/dL/kg) | 1.31 a | 1.14 a | 0.93 b | n.a. | n.a. | n.a. | 0.86 (a) | 0.76 | 0.66 (b) | 0.96 (a) | 0.87 | 0.73 (b) | 1.12 a | 0.87 b | 0.88 b | 0.072 | <0.001 |
| Somatic cell count (103) | 49 | 40 | 93 | n.a. | n.a. | n.a. | 67 | 52 | 73 | 71 | 59 | 204 | 50 b | 41 b | 438 a | 96.3 | 0.01 |
| Feces | |||||||||||||||||
| Mean pH | 7.75 | 7.79 | 7.81 | n.a. | n.a. | n.a. | 7.48 | 7.54 | 7.50 | 7.47 | 7.41 | 7.48 | 7.38 | 7.39 | 7.40 | 0.062 | n.s. |
| Total SCFA (mmol/L) | 52.3 | 57.5 | 56.9 | n.a. | n.a. | n.a. | 89.7 | 93.4 | 82.6 | 78.3 b | 100.7 a | 66.5 b | 88.6 | 86.7 | 82.8 | 8.83 | 0.03 |
| Acetate (%) | 74.9 | 75.6 | 75.8 | n.a. | n.a. | n.a. | 73.8 | 72.5 b | 75.6 a | 75.3 | 74.4 | 74.8 | 74.5 | 75.4 | 73.2 | 1.11 | 0.05 |
| Propionate (%) | 14.5 | 14.2 | 14.1 | n.a. | n.a. | n.a. | 12.7 (b) | 13.7 (a) | 13.7 (a) | 14.0 | 13.5 | 13.9 | 12.4 b | 13.0 | 13.9 a | 0.40 | 0.02 |
| Butyrate (%) | 4.3 | 4.1 | 4.4 | n.a. | n.a. | n.a. | 5.3 | 5.2 | 5.3 | 5.5 | 5.1 | 5.0 | 5.0 | 4.8 | 4.7 | 0.25 | n.s. |
| Valerate (%) | 1.22 | 1.11 | 1.10 | n.a. | n.a. | n.a. | 1.3 a | 1.03 b | 0.99 b | 1.07 | 0.97 | 1.00 | 1.04 | 0.91 | 1.04 | 0.079 | 0.02 |
| Caproate (%) | 0.15 | 0.15 | 0.19 | n.a. | n.a. | n.a. | 2.32 | 3.57 (a) | 0.07 (b) | 0.07 | 1.11 | 0.08 | 3.14 | 1.95 | 2.49 | 1.329 | 0.10 |
| iso-Butyrate (%) | 3.97 | 3.85 | 3.80 | n.a. | n.a. | n.a. | 4.20 | 3.49 | 4.06 | 3.71 (b) | 4.27 | 4.87 (a) | 3.53 | 3.51 | 4.34 | 0.379 | 0.07 |
| iso-Valerate (%) | 0.94 a | 0.90 a | 0.66 b | n.a. | n.a. | n.a. | 0.41 | 0.37 | 0.28 | 0.43 | 0.51 | 0.42 | 0.38 | 0.31 | 0.46 | 0.071 | 0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neubauer, V.; Petri, R.M.; Humer, E.; Kröger, I.; Reisinger, N.; Baumgartner, W.; Wagner, M.; Zebeli, Q. Starch-Rich Diet Induced Rumen Acidosis and Hindgut Dysbiosis in Dairy Cows of Different Lactations. Animals 2020, 10, 1727. https://doi.org/10.3390/ani10101727
Neubauer V, Petri RM, Humer E, Kröger I, Reisinger N, Baumgartner W, Wagner M, Zebeli Q. Starch-Rich Diet Induced Rumen Acidosis and Hindgut Dysbiosis in Dairy Cows of Different Lactations. Animals. 2020; 10(10):1727. https://doi.org/10.3390/ani10101727
Chicago/Turabian StyleNeubauer, Viktoria, Renee M. Petri, Elke Humer, Iris Kröger, Nicole Reisinger, Walter Baumgartner, Martin Wagner, and Qendrim Zebeli. 2020. "Starch-Rich Diet Induced Rumen Acidosis and Hindgut Dysbiosis in Dairy Cows of Different Lactations" Animals 10, no. 10: 1727. https://doi.org/10.3390/ani10101727
APA StyleNeubauer, V., Petri, R. M., Humer, E., Kröger, I., Reisinger, N., Baumgartner, W., Wagner, M., & Zebeli, Q. (2020). Starch-Rich Diet Induced Rumen Acidosis and Hindgut Dysbiosis in Dairy Cows of Different Lactations. Animals, 10(10), 1727. https://doi.org/10.3390/ani10101727





