Histological Analysis, Bioinformatics Profile, and Expression of Methylenetetrahydrofolate Reductase (MTHFR) in Bovine Testes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents, Chemicals, and Instruments
2.2. Animals and Tissue Sample Collection
2.3. Histological Studies of Yellow-Cattle and Yak Testes and Determination of Testicular Index
2.4. Primer Design, Amplification, and MTHFR CDS Sequencing
2.5. Bioinformatics Studies
2.5.1. Analysis and Alignment of DNA and Amino Acids Sequences
2.5.2. MTHFR Protein Secondary and 3D Structure Prediction
2.6. Extraction of Total RNA and Reverse Transcription
2.7. qRT-PCR and MTHFR mRNA Expression Studies in Different Bovine Tissues
2.8. Protein Extraction and Western Blot Analysis
2.9. Statistical Analysis
3. Results
3.1. Histological Evaluation of MTHFR Protein in Yellow-Cattle and Yak Testes and Determination of Testicular Index
3.2. Characterization of the Full Coding Region of Yellow-Cattle MTHFR Gene
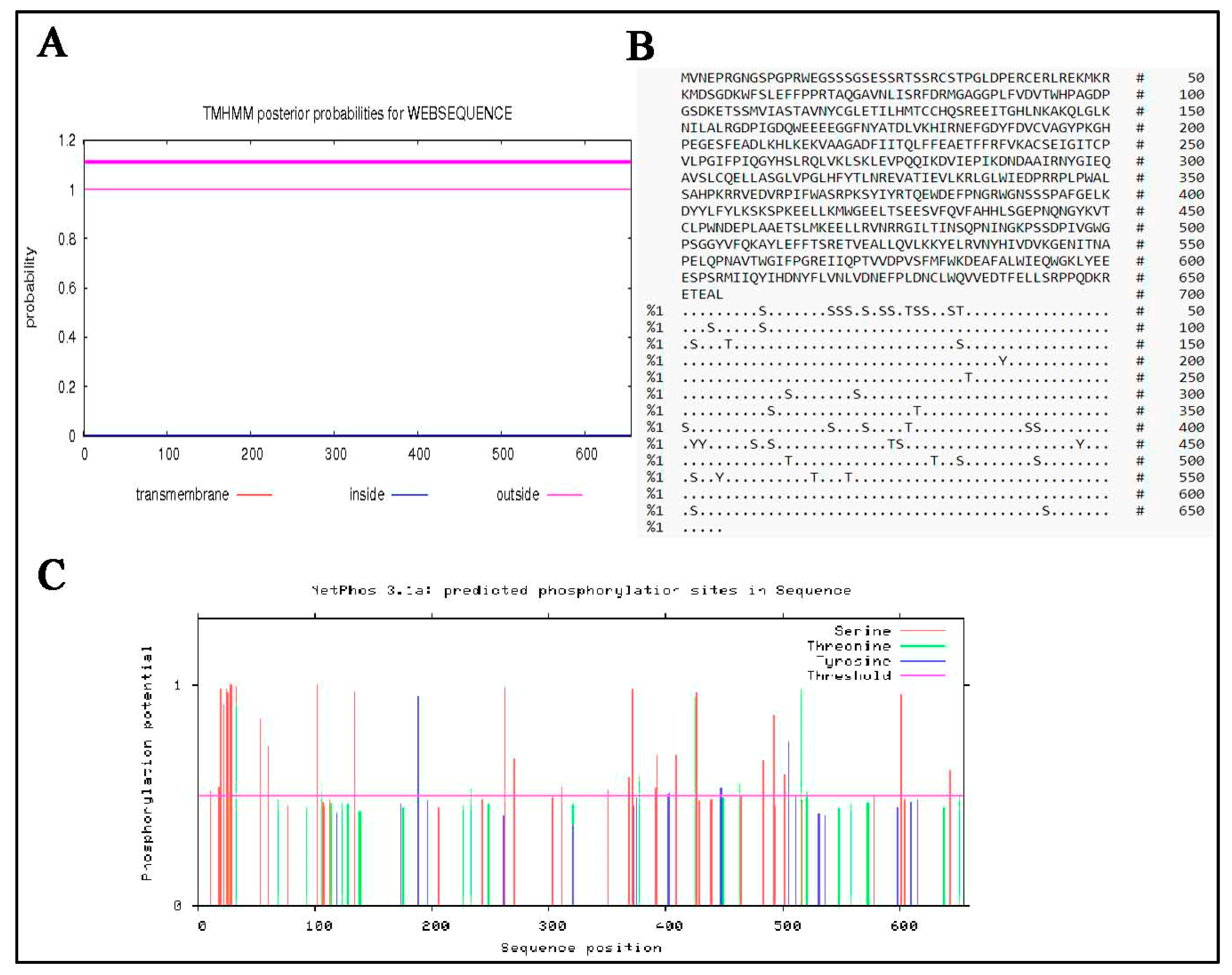
3.3. Molecular Composition of MTHFR Protein
3.4. Multiple Sequence Alignment and Phylogenetic Analysis
3.5. Analysis of the Physical and Chemical Characteristics of Yellow-Cattle MTHFR Protein
3.6. Predicted Secondary and 3D Structure Characteristics of MTHFR Protein
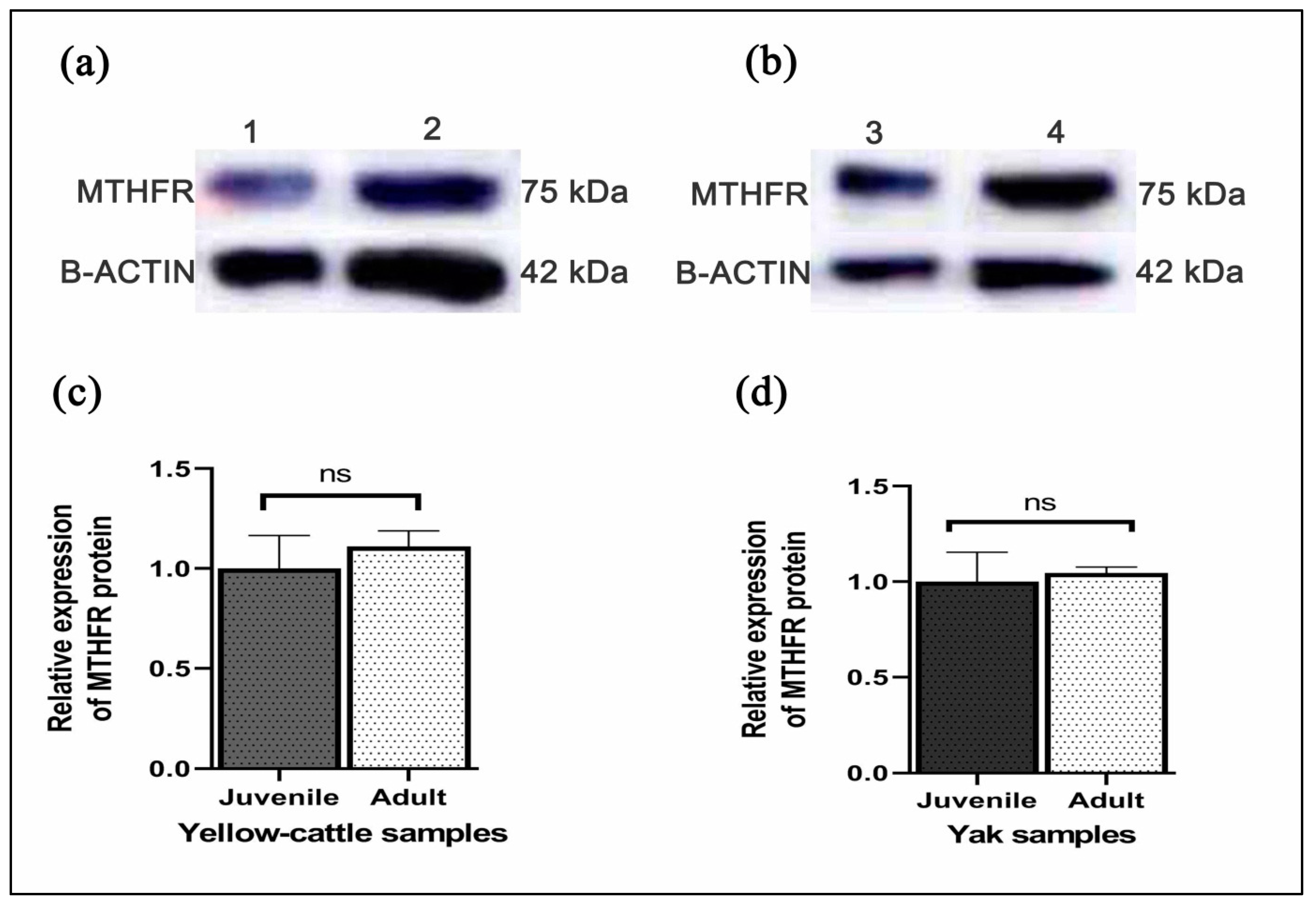
3.7. Expression of MTHFR mRNA and Protein in Yellow-Cattle and Yak Testes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wiener, G.; Han, Y.; Long, R. The Yak, 2nd ed.; Regional Office for Asia and the Pacific, Food and Agriculture Organization of the United Nations: Bangkok, Thailand, 2003. [Google Scholar]
- Zhang, G.; Wang, L.; Chen, H.; Guan, J.; Wu, Y.; Zhao, J.; Luo, Z.; Huang, W.; Zuo, F. Promoter hypermethylation of PIWI/piRNA pathway genes associated with diminished pachytene piRNA production in bovine hybrid male sterility. Epigenetics 2020, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.B.; Han, J.-L.; Wang, G.; Rege, J.E.O.; Hanotte, O. Assessment of cattle genetic introgression into domestic yak populations using mitochondrial and microsatellite DNA markers. Anim. Genet. 2010, 41, 242–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, R.J.; Ding, L.M.; Shang, Z.H.; Guo, X.H. The yak grazing system on the Qinghai-Tibetan plateau and its status. Rangel. J. 2008, 30, 241–246. [Google Scholar] [CrossRef]
- Zhang, R.C. Interspecies hybridization between Yak, bos taurus and bos indicus and reproduction of the hybrids. In Recent Advances in Yak Reproduction; Zhao, X.X., Zhang, R.C., Eds.; IVIS: Ithaca, NY, USA, 2000. [Google Scholar]
- Krausz, C.; Riera-Escamilla, A. Genetics of male infertility. Nat. Rev. Urol. 2018, 15, 369–384. [Google Scholar] [CrossRef]
- Wang, S.; Pan, Z.; Zhang, Q.; Xie, Z.; Liu, H.; Li, Q. Differential mRNA Expression and Promoter Methylation Status of SYCP3 Gene in Testes of Yaks and Cattle-Yaks. Reprod. Domest. Anim. 2012, 47, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, Q.; Zhang, Q.; Qu, X.; Dong, L.; Yang-zom, C.; Xie, Z.; Liu, H. Analysis of IGF2 mRNA expression and its methylation status between cattle yaks and their parents. Prog. Nat. Sci. 2009, 19, 1063–1069. [Google Scholar] [CrossRef]
- Johnson, L.; Varner, D.D.; Roberts, M.E.; Smith, T.L.; Keillor, G.E.; Scrutchfield, W.L. Efficiency of spermatogenesis: A comparative approach. Anim. Reprod. Sci. 2000, 60, 471–480. [Google Scholar] [CrossRef]
- Lou, Y.N.; Liu, W.J.; Wang, C.L.; Huang, L.; Jin, S.Y.; Lin, Y.Q.; Zheng, Y.C. Histological evaluation and Prdm9 expression level in the testis of sterile male cattle-yaks. Livest. Sci. 2014, 160, 208–213. [Google Scholar] [CrossRef]
- Datar, J.; Regassa, A.; Kim, W.-K.; Taylor, C.G.; Zahradka, P.; Suh, M. Lipid Metabolism is Closely Associated with Normal Testicular Growth Based on Global Transcriptome Profiles in Normal and Underdeveloped Testis of Obese Zucker (fa/fa) Rats. Lipids 2017, 52, 951–960. [Google Scholar] [CrossRef]
- Plant, T.M.; Zeleznik, A.J. Knobil and Neill’s Physiology of Reproduction, 3rd ed.; Neill, J.D., Knobil, E., Eds.; Academic Press: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Mobasheri, M.B.; Babatunde, K.A. Testicular miRNAs in relation to spermatogenesis, spermatogonial stem cells and cancer/testis genes. Sci. Afr. 2019, 3, e00067. [Google Scholar] [CrossRef]
- Welsh, M.; Saunders, P.T.K.; Atanassova, N.; Sharpe, R.M.; Smith, L.B. Androgen action via testicular peritubular myoid cells is essential for male fertility. FASEB J. 2009, 23, 4218–4230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Gendt, K.; Swinnen, J.V.; Saunders, P.T.K.; Schoonjans, L.; Dewerchin, M.; Devos, A.; Tan, K.; Atanassova, N.; Claessens, F.; Lécureuil, C.; et al. A Sertoli Cell-Selective Knockout of the Androgen Receptor Causes Spermatogenic Arrest in Meiosis. Proc. Nat. Acad. Sci. USA 2004, 101, 1327–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šerý, O.; Šrámková, T.; Klempová, J.; Šťastný, F.; Lochman, J.; Khan, N.A. The relationship between the C677T polymorphism of the MTHFR gene and serum levels of luteinizing hormone in males with erectile dysfunction. Neuro Endocrinol. Lett. 2012, 33, 7. [Google Scholar]
- Hyde, Z.; Norman, P.E.; Flicker, L.; Hankey, G.J.; McCaul, K.A.; Almeida, O.P.; Chubb, S.A.P.; Yeap, B.B. Elevated LH predicts ischaemic heart disease events in older men: The Health in Men Study. Eur. J. Endocrinol. 2011, 164, 569–577. [Google Scholar] [CrossRef] [Green Version]
- Gong, M.; Dong, W.; He, T.; Shi, Z.; Huang, G.; Ren, R.; Huang, S.; Qiu, S.; Yuan, R. MTHFR 677C>T Polymorphism Increases the Male Infertility Risk: A Meta-Analysis Involving 26 Studies. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Hoffman, A.; Taleski, G.; Qian, H.; Wasek, B.; Arning, E.; Bottiglieri, T.; Sontag, J.-M.; Sontag, E. Methylenetetrahydrofolate Reductase Deficiency Deregulates Regional Brain Amyloid-β Protein Precursor Expression and Phosphorylation Levels. J. Alzheimers Dis. 2018, 64, 223–237. [Google Scholar] [CrossRef]
- Kelly, T.L.J.; Neaga, O.R.; Schwahn, B.C.; Rozen, R.; Trasler, J.M. Infertility in 5,10-Methylenetetrahydrofolate Reductase (MTHFR)-deficient male mice is partially alleviated by lifetime dietary betaine supplementation1. Biol. Reprod. 2005, 72, 667–677. [Google Scholar] [CrossRef] [Green Version]
- Rajender, S.; Avery, K.; Agarwal, A. Epigenetics, spermatogenesis and male infertility. Mutat. Res. Rev. Mutat. Res. 2011, 727, 62–71. [Google Scholar] [CrossRef]
- Desai, M.; Chauhan, J.B. Computational analysis for the determination of deleterious nsSNPs in human MTHFR gene. Comput. Biol. Chem. 2018, 74, 20–30. [Google Scholar] [CrossRef]
- Khazamipour, N.; Noruzinia, M.; Fatehmanesh, P.; Keyhanee, M.; Pujol, P. MTHFR promoter hypermethylation in testicular biopsies of patients with non-obstructive azoospermia: The role of epigenetics in male infertility. Hum. Reprod. 2009, 24, 2361–2364. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Shen, O.; Qin, Y.; Lu, J.; Niu, X.; Zhou, Z.; Lu, C.; Xia, Y.; Wang, S.; Wang, X. Methylenetetrahydrofolate reductase C677T polymorphism and the risk of male infertility: A metaanalysis. Int. J. Androl. 2011, 35, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.J.; Costa, P.; Vaz, B.; Carvalho, F.; Fernandes, S.; Barros, A.; Sousa, M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol. Hum. Reprod. 2008, 14, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Karaplis, A.C.; Ackerman, S.L.; Pogribny, I.P.; Melnyk, S.; Lussier-Cacan, S.; Pai, A.; John, S.W.M.; Smith, R.S.; Bottiglieri, T.; et al. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum. Mol. Genet. 2001, 10, 433–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goyette, P.; Sumner, J.S.; Milos, R.; Duncan, A.M.V.; Rosenblatt, D.S.; Matthews, R.G.; Rozen, R. Human methylenetetrahydrofolate reductase: Isolation of cDNA, mapping and mutation identification. Nat. Genet. 1994, 7, 195–200. [Google Scholar] [CrossRef]
- Xu, A.; Hua, Y.; Zhang, J.; Chen, W.; Zhao, K.; Xi, W.; Wang, H.; Fang, J.; Su, S.; Tang, M.; et al. Abnormal Hypermethylation of the VDAC2 Promoter is a Potential Cause of Idiopathic Asthenospermia in Men. Sci. Rep. 2016, 6, 37836. [Google Scholar] [CrossRef] [Green Version]
- Bailey, L.B.; Gregory, J.F. Polymorphisms of Methylenetetrahydrofolate reductase and other enzymes: Metabolic significance, risks and impact on folate requirement. J. Nutr. 1999, 129, 919–922. [Google Scholar] [CrossRef]
- Shen, H.; Spitz, M.R.; Wang, L.-E.; Hong, W.K.; Wei, Q. Polymorphisms of Methylene-tetrahydrofolate Reductase and Risk of Lung Cancer: A Case-Control Study. Cancer Epidemiol. Prev. Biomark. 2001, 10, 397–401. [Google Scholar]
- Zhang, Q.; Gong, J.; Wang, X.; Wu, X.; Li, Y.; Ma, Y.; Zhang, Y.; Zhao, X. Molecular Cloning, Bioinformatics Analysis and Expression of Insulin-Like Growth Factor 2 from Tianzhu White Yak, Bos grunniens. Int. J. Mol. Sci. 2014, 14, 504–524. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.M.R.; Guo, D.; Hodges, R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: Correlation of predicted surface residues with antigenicity and x-ray-derived accessible sites. Biochemistry 1986, 25, 5425–5432. [Google Scholar] [CrossRef]
- Gaughan, D.J.; Barbaux, S.; Kluijtmans, L.A.J.; Whitehead, A.S. The human and mouse methylenetetrahydrofolate reductase (MTHFR) genes: Genomic organization, mRNA structure and linkage to the CLCN6 gene. Gene 2000, 257, 279–289. [Google Scholar] [CrossRef]
- Lodhi, L.A.; Qureshi, Z.I.; Ali, F. The bovine testis—I: Pre and post natal development. Pak. J. Biol. Sci. 2000, 3, 1691–1696. [Google Scholar]
- Curtis, S.K.; Amann, R.P. Testicular Development and Establishment of Spermatogenesis in Holstein Bulls. J. Anim. Sci. 1981, 53, 1645–1657. [Google Scholar] [CrossRef] [PubMed]
- Homberger, A.; Linnebank, M.; Winter, C.; Willenbring, H.; Marquardt, T.; Harms, E.; Koch, H.G. Genomic structure and transcript variants of the human methylenetetrahydrofolate reductase gene. Eur. J. Hum. Genet. 2000, 8, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Frottin, F.; Martinez, A.; Peynot, P.; Mitra, S.; Holz, R.C.; Giglione, C.; Meinnel, T. The proteomics of N-terminal methionine cleavage. Mol. Cell Proteom. 2006, 5, 2336–2349. [Google Scholar] [CrossRef] [Green Version]
- Bradshaw, R.A.; Brickey, W.W.; Walker, K.W. N-Terminal processing: The methionine aminopeptidase and N-acetyl transferase families. Trends Biochem. Sci. 1998, 523, 263–267. [Google Scholar] [CrossRef]
- Giglione, C.; Boularot, A.; Meinnel, T. Protein N-terminal methionine excision. CMLS Cell. Mol. Life Sci. 2004, 61. [Google Scholar] [CrossRef]
- Scheiner, S.; Kar, T.; Pattanayak, J. Comparison of Various Types of Hydrogen Bonds Involving Aromatic Amino Acids. J. Am. Chem. Soc. 2002, 124, 13257–13264. [Google Scholar] [CrossRef]
- Xu, Y.-Y.; Yao, L.-X.; Shen, H.-B. Bioimage-based protein subcellular location prediction: A comprehensive review. Front. Comput. Sci. 2017, 12, 26–39. [Google Scholar] [CrossRef]
- Hung, M.-C.; Link, W. Protein localization in disease and therapy. J. Cell Sci. 2011, 124, 3381–3392. [Google Scholar] [CrossRef] [Green Version]
- Komor, A.C.; Schneider, C.J.; Weidmann, A.G.; Barton, J.K. Cell-Selective Biological Activity of Rhodium Metalloinsertors Correlates with Subcellular Localization. J. Am. Chem. Soc. 2012, 134, 19223–19233. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Byun, K.; Hong, W.; Chuang, H.-Y.; Pack, C.-G.; Bayarsaikhan, E.; Paek, S.H.; Kim, H.; Shin, H.Y.; Ideker, T.; et al. Proteome-wide discovery of mislocated proteins in cancer. Genome Res. 2013, 23, 1283–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Hu, J. Mislocalization-related disease gene discovery using gene expression based computational protein localization prediction. Methods 2016, 93, 119–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, S.T.; Lau, L.-F. Tyrosine phosphorylation. In Basic Neurochemistry; Elsevier: Amsterdam, The Netherlands, 2012; pp. 493–513. [Google Scholar]
- Plattner, F.; Bibb, J.A. Serine and threonine phosphorylation. In Basic Neurochemistry; Elsevier: Amsterdam, The Netherlands, 2012; pp. 467–492. [Google Scholar]
- Fodje, M.N.; Al-Karadaghi, S. Occurrence, conformational features and amino acid propensities for the π-helix. Protein Eng. Des. Sel. 2002, 15, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, H.; L’Hernault, S.W. Spermatogenesis. Curr. Biol. 2017, 27, R988–R994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chocu, S.; Calvel, P.; Rolland, A.D.; Pineau, C. Spermatogenesis in mammals: Proteomic insights. Syst. Biol. Reprod. Med. 2012, 58, 179–190. [Google Scholar] [CrossRef]
- Liu, P.; Yu, S.; Cui, Y.; He, J.; Zhang, Q.; Sun, J.; Huang, Y.; Yang, X.; Cao, M.; Liao, B.; et al. Regulation by Hsp27/P53 in testis development and sperm apoptosis of male cattle (cattle-yak and yak). J. Cell Physiol. 2019, 234, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Daubner, S.C.; Matthews, R.G. Purification and properties of methylenetetrahydrofolate reductase from pig liver. J. Biol. Chem. 1982, 257, 140–145. [Google Scholar]






| Animal Species | Number of Animals (Testes) | Age Group | Age (Year) | Source (All in China) | Application |
|---|---|---|---|---|---|
| Yellow-cattle | 3 (6) | Juvenile | 1–2 | Wuwei city | Histology and molecular biology studies |
| 3 (6) | Adult | 3 | |||
| Yak | 3 (6) | Juvenile | 1 | Linxia city | |
| 4 (8) | Adult | 4 |
| Primer Name | Sequence (5′ to 3′) | Tm (°C) | Product Size (bp) | Accession No |
|---|---|---|---|---|
| MTHFR F-1 | TCGTGGATGTGAAGGGTGAA | 55.40 | 187 | NM_001011685.1 |
| MTHFR R-1 | TCCTCCTCATACAGCTTGCC | 57.45 | ||
| MTHFR F-2 | GGAGAGAGCTTTGAGGCTGA | 57.45 | 185 | NM_001011685.1 |
| MTHFR R-2 | AGGGAGTGGTAGCCCTGAAT | 57.45 | ||
| β-Actin F | GCAATGAGCGGTTCC | 56.00 | 141 | XM_005887322.2 |
| β-Actin R | CCGTGTTGGCGTAGAG | 56.00 |
| Animal Specie | Age Group | Age (Years) | Body Weight (kg) at Slaughter | Testicular Weight (g) | Testicular Index (g/kg × 100) |
|---|---|---|---|---|---|
| Yellow-cattle | Adult | 3 | 569.67 ± 9.00 | 310.53 ± 5.85 | 55.01 ± 0.01 a |
| Juvenile | 1–2 | 275.47 ± 7.64 | 117.21 ± 8.17 | 42.52 ± 0.02 b | |
| Yak | Adult | 4 | 396.19 ± 9.43 | 152.28 ± 8.43 | 38.42 ± 0.01 c |
| Juvenile | 1 | 292.57 ± 5.59 | 102.59 ± 4.41 | 35.06 ± 0.01 d |
| Animal Species MTHFR | Accession Number | No. of Amino Acid Residue | Total Score (%) | Identity (%) | Query Cover (%) | E-Value | pI | Molecular Weight |
|---|---|---|---|---|---|---|---|---|
| Bos taurus | NP_001011685.1 | 655 | 1357 | 100.00 | 100 | 0.0 | 5.30 | 74.49 |
| Bos mutus | XP_005889249.1 | 696 | 1349 | 99.24 | 100 | 0.0 | 5.44 | 78.80 |
| Bos indicus | XP_019831767.1 | 696 | 1347 | 99.08 | 100 | 0.0 | 5.44 | 78.78 |
| Bison bison | XP_010848171.1 | 695 | 1348 | 99.24 | 100 | 0.0 | 5.44 | 78.72 |
| Capra hircus | XP_005690729.2 | 799 | 1342 | 98.63 | 100 | 0.0 | 5.78 | 89.66 |
| Ovis aries | XP_027831433.1 | 655 | 1342 | 98.63 | 100 | 0.0 | 5.18 | 74.52 |
| Bubalus bubalis | XP_006076816.2 | 655 | 1341 | 98.47 | 100 | 0.0 | 5.22 | 74.58 |
| Camelus bactrianus | XP_010955831.1 | 655 | 1254 | 93.58 | 99 | 0.0 | 5.23 | 74.67 |
| Canis lupus familiaris | XP_005618050.1 | 656 | 1215 | 91.02 | 100 | 0.0 | 5.22 | 74.45 |
| Equus caballus | XP_005607580.1 | 655 | 1231 | 92.20 | 99 | 0.0 | 5.11 | 74.45 |
| Sus scrofa, | XP_020951001.1 | 700 | 1248 | 89.02 | 99 | 0.0 | 5.39 | 78.94 |
| Homo sapiens | CAB41971.1 | 679 | 1209 | 89.92 | 99 | 0.0 | 5.30 | 76.88 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afedo, S.Y.; Cui, Y.; Yu, S.; Liao, B.; Zhao, Z.; Li, H.; Zhang, H.; Zou, S.; Li, D.H.; Zhang, P. Histological Analysis, Bioinformatics Profile, and Expression of Methylenetetrahydrofolate Reductase (MTHFR) in Bovine Testes. Animals 2020, 10, 1731. https://doi.org/10.3390/ani10101731
Afedo SY, Cui Y, Yu S, Liao B, Zhao Z, Li H, Zhang H, Zou S, Li DH, Zhang P. Histological Analysis, Bioinformatics Profile, and Expression of Methylenetetrahydrofolate Reductase (MTHFR) in Bovine Testes. Animals. 2020; 10(10):1731. https://doi.org/10.3390/ani10101731
Chicago/Turabian StyleAfedo, Seth Yaw, Yan Cui, Sijiu Yu, Bo Liao, Zihan Zhao, Hui Li, Huizhu Zhang, Shengnan Zou, De Hong Li, and Peng Zhang. 2020. "Histological Analysis, Bioinformatics Profile, and Expression of Methylenetetrahydrofolate Reductase (MTHFR) in Bovine Testes" Animals 10, no. 10: 1731. https://doi.org/10.3390/ani10101731
APA StyleAfedo, S. Y., Cui, Y., Yu, S., Liao, B., Zhao, Z., Li, H., Zhang, H., Zou, S., Li, D. H., & Zhang, P. (2020). Histological Analysis, Bioinformatics Profile, and Expression of Methylenetetrahydrofolate Reductase (MTHFR) in Bovine Testes. Animals, 10(10), 1731. https://doi.org/10.3390/ani10101731





