Influence of Parental Fatty Acid Desaturase 2 (fads2) Expression and Diet on Gilthead Seabream (Sparus aurata) Offspring fads2 Expression during Ontogenesis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Broodstock fads2 Expression
2.3. Effect of the Broodstock Diet
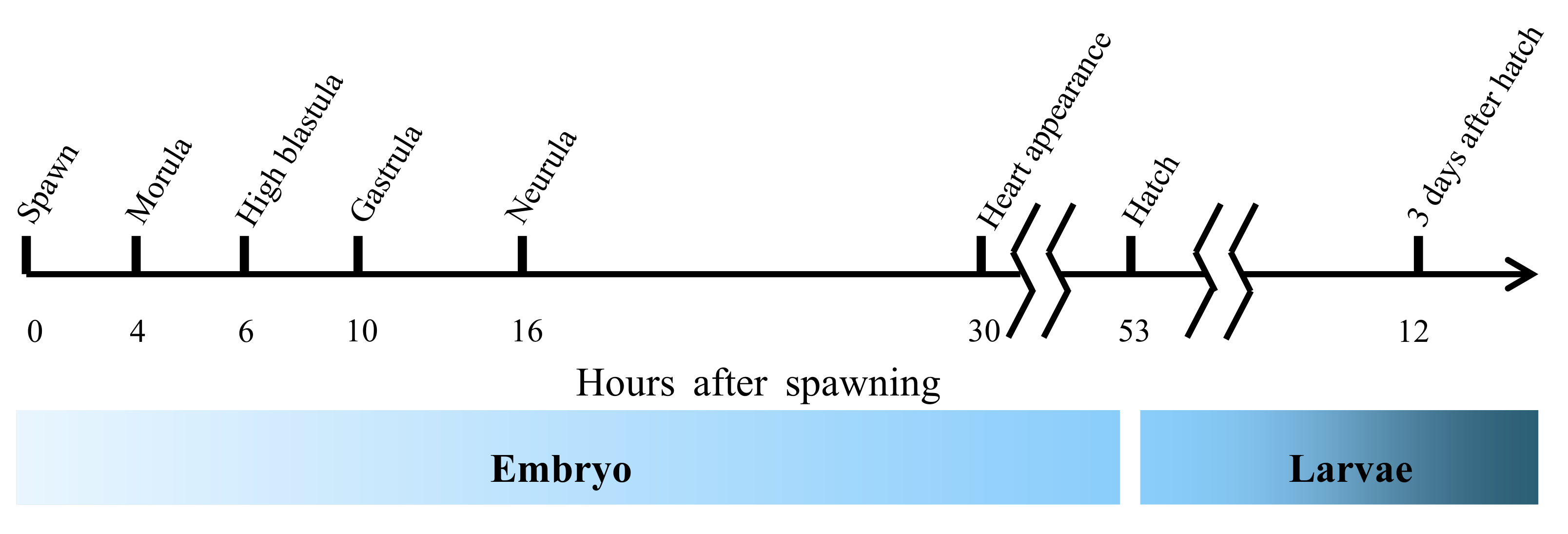
2.4. Sampling during Offspring Ontogenesis
2.5. RNA Extraction and Digital PCR
2.6. Lipid and Fatty Acid Analysis
2.7. Data Analysis
3. Results
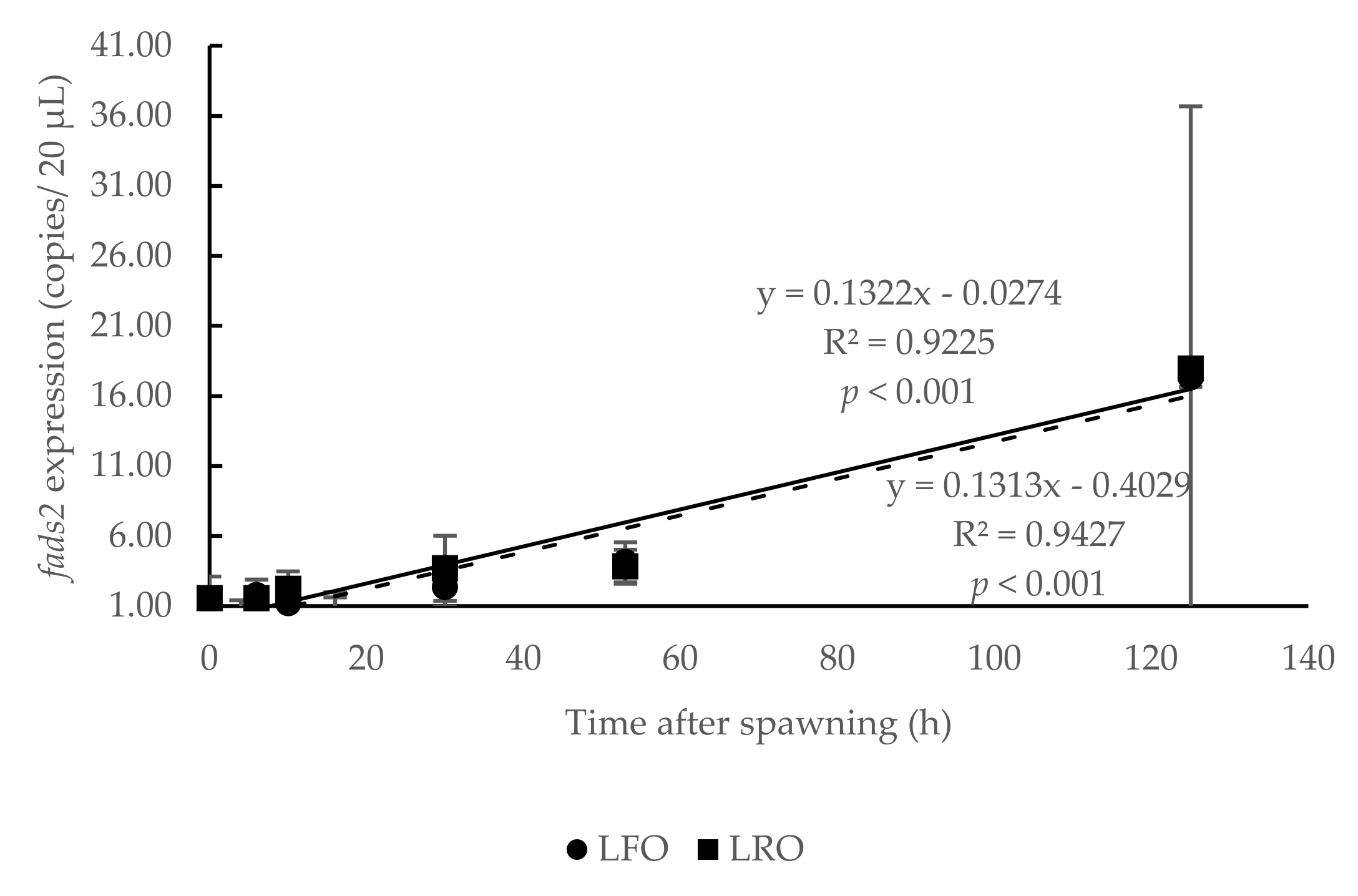
3.1. Temporal Expression of fads2 during Offspring Ontogenesis
3.2. Influence of Parental fads2 Expression in Offspring fads2 Expression during Ontogenesis
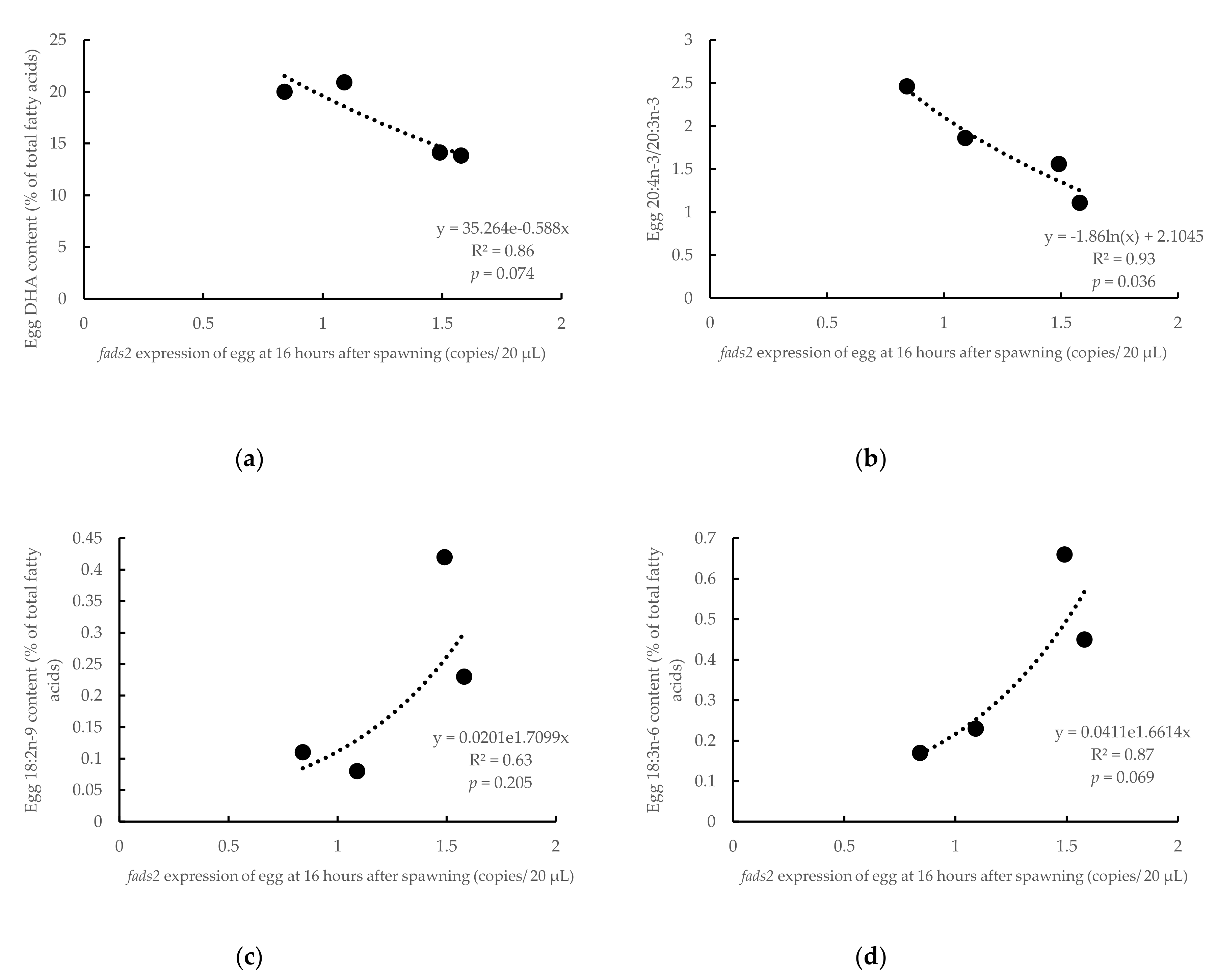
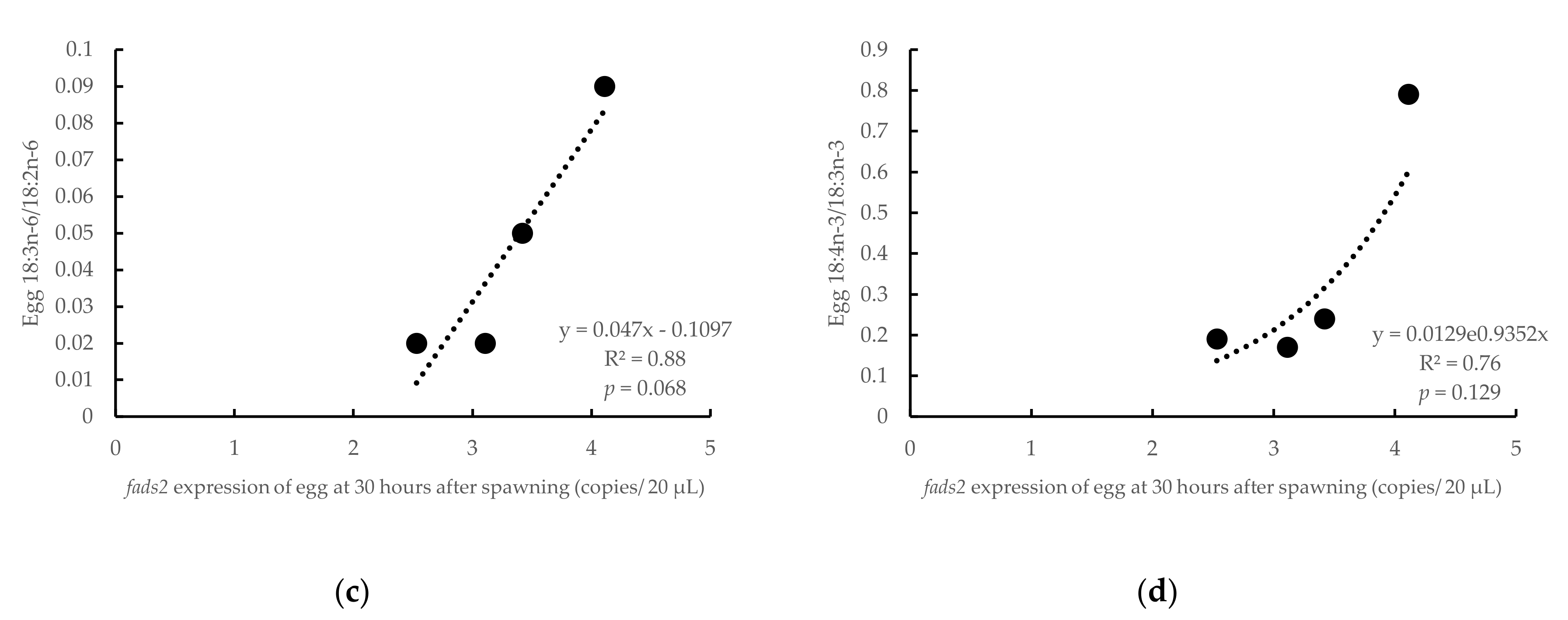
3.3. Influence of Parental fads2 Expression in Egg Fatty Acid Profiles
3.4. Influence of Broodstock Diet in Offspring fads2 Expression during Ontogenesis
4. Discussion
4.1. Changes of fads2 mRNA in Gilthead Seabream Eggs during Ontogeny
4.2. Influence of Parental fads2 Expression in Offspring fads2 Expression during Ontogenesis
4.3. Influence of Broodstock Diet in Offspring fads2 Expression during Ontogenesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LC-PUFAs | long chain polyunsaturated fatty acids |
| DHA | docosahexaenoic acid |
| EPA | eicosapentaenoic acid |
| ARA | arachidonic acid |
| FO | fish oil |
| fads | fatty acyl desaturase |
| FM | fishmeal |
| hps | hour post spawning |
| SFA | saturated fatty acid |
| MUFA | monounsaturated fatty acid |
| S.D. | standard deviation |
References
- Calder, P.C. Mechanisms of Action of (n-3) Fatty Acids. J. Nutr. 2012, 142, 592S–599S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, T. Importance of Docosahexaenoic Acid in Marine Larval Fish. J. World Aquac. Soc. 1993, 24, 152–161. [Google Scholar] [CrossRef]
- Wassall, S.R.; Stillwell, W. Docosahexaenoic acid domains: The ultimate non-raft membrane domain. Chem. Phys. Lipids 2008, 153, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Gill, I.; Valivety, R. Polyunsaturated fatty acids, part 1: Occurrence, biological activities and applications. Trends Biotechnol. 1997, 15, 401–409. [Google Scholar] [CrossRef]
- Peng, S.; Yue, Y.; Gao, Q.; Shi, Z.; Yin, F.; Wang, J. Influence of dietary n-3 LC-PUFA on growth, nutritional composition and immune function in marine fish Sebastiscus marmoratus. Chin. J. Oceanol. Limnol. 2014, 32, 1000–1008. [Google Scholar] [CrossRef]
- Izquierdo, M.; Koven, W. Digestive development and nutrient requirements. In Larval Fish Nutrition; John Wiley & Sons, Inc.: Chichester, UK, 2011; pp. 47–81. [Google Scholar]
- Menoyo, D.; Izquierdo, M.S.; Robaina, L.; Ginés, R.; Lopez-Bote, C.J.; Bautista, J.M. Adaptation of lipid metabolism, tissue composition and flesh quality in gilthead sea bream (Sparus aurata) to the replacement of dietary fish oil by linseed and soyabean oils. Br. J. Nutr. 2004, 92, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Torrecillas, S.; Robaina, L.; Caballero, M.; Montero, D.; Calandra, G.; Mompel, D.; Karalazos, V.; Kaushik, S.; Izquierdo, M. Combined replacement of fishmeal and fish oil in European sea bass (Dicentrarchus labrax): Production performance, tissue composition and liver morphology. Aquaculture 2017, 474, 101–112. [Google Scholar] [CrossRef]
- Carvalho, M.; Montero, D.; Rosenlund, G.; Fontanillas, R.; Ginés, R.; Izquierdo, M. Effective complete replacement of fish oil by combining poultry and microalgae oils in practical diets for gilthead sea bream (Sparus aurata) fingerlings. Aquaculture 2020, 529, 735696. [Google Scholar] [CrossRef]
- Izquierdo, M.; Turkmen, S.; Montero, D.; Zamorano, M.; Afonso, J.; Karalazos, V.; Fernández-Palacios, H. Nutritional programming through broodstock diets to improve utilization of very low fishmeal and fish oil diets in gilthead sea bream. Aquaculture 2015, 449, 18–26. [Google Scholar] [CrossRef]
- Pereira, S.L.; Leonard, A.E.; Mukerji, P. Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins Leukot. Essent. Fat. Acids 2003, 68, 97–106. [Google Scholar] [CrossRef]
- Lopes-Marques, M.; Kabeya, N.; Qian, Y.; Ruivo, R.; Santos, M.M.; Venkatesh, B.; Tocher, D.R.; Castro, L.F.C.; Monroig, Ó. Retention of fatty acyl desaturase 1 (fads1) in Elopomorpha and Cyclostomata provides novel insights into the evolution of long-chain polyunsaturated fatty acid biosynthesis in vertebrates. BMC Evol. Biol. 2018, 18, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprecher, H. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2000, 1486, 219–231. [Google Scholar] [CrossRef]
- Dong, X.; Tan, P.; Cai, Z.; Xu, H.; Li, J.; Ren, W.; Xu, H.; Zuo, R.; Zhou, J.; Mai, K.; et al. Regulation of FADS2 transcription by SREBP-1 and PPAR-α influences LC-PUFA biosynthesis in fish. Sci. Rep. 2017, 7, 40024. [Google Scholar] [CrossRef] [PubMed]
- Tocher, D.R.; Ghioni, C. Fatty acid metabolism in marine fish: Low activity of fatty acyl Δ5 desaturation in gilthead sea bream (Sparus aurata) cells. Lipids 1999, 34, 433–440. [Google Scholar] [CrossRef]
- Vagner, M.; Santigosa, E. Characterization and modulation of gene expression and enzymatic activity of delta-6 desaturase in teleosts: A review. Aquaculture 2011, 315, 131–143. [Google Scholar] [CrossRef]
- Tocher, D.R.; Zheng, X.; Schlechtriem, C.; Hastings, N.; Dick, J.R.; Teale, A.J. Highly unsaturated fatty acid synthesis in marine fish: Cloning, functional characterization, and nutritional regulation of fatty acyl Δ6 desaturase of Atlantic cod (Gadus morhua L.). Lipids 2006, 41, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Sargent, J.R.; Tocher, D.R.; Bell, J.G. The lipids. In Fish Nutrition, 3rd ed.; Elsevier: San Diego, CA, USA, 2003; pp. 181–257. [Google Scholar]
- Izquierdo, M.S. Essential fatty acid requirements of cultured marine fish larvae. Aquac. Nutr. 1996, 2, 183–191. [Google Scholar] [CrossRef]
- Kabeya, N.; Yoshizaki, G.; Tocher, D.R.; Monroig, Ó. Diversification of Fads2 in finfish species: Implications for aquaculture. In Investigación y Desarrollo en Nutrición Acuícola; Cruz-Suarez, L.E., Ricque-Marie, D., Tapia-Salazar, M., Nieto-López, M.G., Villarreal-Cavazos, D.A., Gamboa-Delgado, J., López Acuña, L.M., Galaviz-Espinoza, M., Eds.; Universidad Autónoma de Nuevo León San Nicolás de los Garza: Nuevo León, México, 2017; pp. 338–362. ISBN 978-607-27-0822-8. [Google Scholar]
- Monroig, Ó.; Li, Y.; Tocher, D.R. Delta-8 desaturation activity varies among fatty acyl desaturases of teleost fish: High activity in delta-6 desaturases of marine species. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2011, 159, 206–213. [Google Scholar] [CrossRef]
- Ferosekhan, S.; Xu, H.; Turkmen, S.; Gómez, A.; Afonso, J.M.; Fontanillas, R.; Rosenlund, G.; Kaushik, S.; Izquierdo, M. Reproductive performance of gilthead seabream (Sparus aurata) broodstock showing different expression of fatty acyl desaturase 2 and fed two dietary fatty acid profiles. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Ferosekhan, S.; Turkmen, S.; Xu, H.; Afonso, J.M.; Zamorano, M.J.; Kaushik, S.J.; Izquierdo, M. The Relationship between the Expression of Fatty Acyl Desaturase 2 (fads2) Gene in Peripheral Blood Cells (PBCs) and Liver in Gilthead Seabream, Sparus aurata Broodstock Fed a Low n-3 LC-PUFA Diet. Life 2020, 10, 117. [Google Scholar] [CrossRef]
- Bell, J.G.; Henderson, R.J.; Tocher, D.R.; McGhee, F.; Dick, J.R.; Porter, A.; Smullen, R.P.; Sargent, J.R. Substituting Fish Oil with Crude Palm Oil in the Diet of Atlantic Salmon (Salmo salar) Affects Muscle Fatty Acid Composition and Hepatic Fatty Acid Metabolism. J. Nutr. 2002, 132, 222–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, M.K.; Collins, R.O.; Tocher, D.R.; James, M.J.; Turchini, G.M. Nutritional regulation of long-chain PUFA biosynthetic genes in rainbow trout (Oncorhynchus mykiss). Br. J. Nutr. 2016, 115, 1721–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Dong, X.; Ai, Q.; Mai, K.; Xu, W.; Zhang, Y.; Zuo, R. Regulation of Tissue LC-PUFA Contents, Δ6 Fatty Acyl Desaturase (FADS2) Gene Expression and the Methylation of the Putative FADS2 Gene Promoter by Different Dietary Fatty Acid Profiles in Japanese Seabass (Lateolabrax japonicus). PLoS ONE 2014, 9, e87726. [Google Scholar] [CrossRef] [PubMed]
- Turkmen, S.; Perera, E.; Zamorano, M.J.; Simó-Mirabet, P.; Xu, H.; Pérez-Sánchez, J.; Izquierdo, M. Effects of Dietary Lipid Composition and Fatty Acid Desaturase 2 Expression in Broodstock Gilthead Sea Bream on Lipid Metabolism-Related Genes and Methylation of the fads2 Gene Promoter in Their Offspring. Int. J. Mol. Sci. 2019, 20, 6250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turkmen, S.; Zamorano, M.J.; Fernández-Palacios, H.; Hernández-Cruz, C.M.; Montero, D.; Robaina, L.; Izquierdo, M. Parental nutritional programming and a reminder during juvenile stage affect growth, lipid metabolism and utilisation in later developmental stages of a marine teleost, the gilthead sea bream (Sparus aurata). Br. J. Nutr. 2017, 118, 500–512. [Google Scholar] [CrossRef] [Green Version]
- Lazzarotto, V.; Corraze, G.; Larroquet, L.; Mazurais, D.; Médale, F. Does broodstock nutritional history affect the response of progeny to different first-feeding diets? A whole-body transcriptomic study of rainbow trout alevins. Br. J. Nutr. 2016, 115, 2079–2092. [Google Scholar] [CrossRef] [Green Version]
- Lazzarotto, V.; Corraze, G.; Leprévost, A.; Quillet, E.; Dupont-Nivet, M.; Médale, F. Three-Year Breeding Cycle of Rainbow Trout (Oncorhynchus mykiss) Fed a Plant-Based Diet, Totally Free of Marine Resources: Consequences for Reproduction, Fatty Acid Composition and Progeny Survival. PLoS ONE 2015, 10, e0117609. [Google Scholar] [CrossRef] [Green Version]
- Monroig, Ó.; Rotllant, J.; Sánchez, E.; Cerdá-Reverter, J.M.; Tocher, D.R. Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2009, 1791, 1093–1101. [Google Scholar] [CrossRef] [Green Version]
- Tanomman, S.; Ketudat-Cairns, M.; Jangprai, A.; Boonanuntanasarn, S. Characterization of fatty acid delta-6 desaturase gene in Nile tilapia and heterogenous expression in Saccharomyces cerevisiae. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2013, 166, 148–156. [Google Scholar] [CrossRef]
- Torres, M.; Navarro, J.; Varó, I.; Agulleiro, M.; Morais, S.; Monroig, Ó.; Hontoria, F. Expression of genes related to long-chain (C18–22) and very long-chain (>C24) fatty acid biosynthesis in gilthead seabream (Sparus aurata) and Senegalese sole (Solea senegalensis) larvae: Investigating early ontogeny and nutritional regulation. Aquaculture 2020, 520, 734949. [Google Scholar] [CrossRef]
- Turkmen, S.; Hernández-Cruz, C.M.; Zamorano, M.J.; Fernández-Palacios, H.; Montero, D.; Afonso, J.M.; Izquierdo, M. Long-chain PUFA profiles in parental diets induce long-term effects on growth, fatty acid profiles, expression of fatty acid desaturase 2 and selected immune system-related genes in the offspring of gilthead seabream. Br. J. Nutr. 2019, 122, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Kamacı, H.O.; Saka, Ş.; Fırat, K. The Cleavage and Embryonic Phase of Gilthead Sea Bream (Sparus aurata Linnaeus, 1758) Eggs. EgeJFAS 2005, 22, 205–209. [Google Scholar]
- Folch, J.; Lees, M.; Stanley, G.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [PubMed]
- Christie, W. Equivalent chain-lengths of methyl ester derivatives of fatty acids on gas chromatography A reappraisal. J. Chromatogr. A 1988, 447, 305–314. [Google Scholar] [CrossRef]
- Takeuchi, T.; Toyota, M.; Satoh, S.; Watanabe, T. Requirement of juvenile red seabream Pagrus major for eicosapentaenoic and docosahexaenoic acids. Nippon Suisan Gakkaishi 1990, 56, 1263–1269. [Google Scholar] [CrossRef]
- Vergauwen, L.; Cavallin, J.E.; Ankley, G.T.; Bars, C.; Gabriëls, I.J.; Michiels, E.D.; Fitzpatrick, K.R.; Periz-Stanacev, J.; Randolph, E.C.; Robinson, S.L.; et al. Gene transcription ontogeny of hypothalamic-pituitary-thyroid axis development in early-life stage fathead minnow and zebrafish. Gen. Comp. Endocrinol. 2018, 266, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.T.; Pasquier, J.; Bouleau, A.; Nguyen, T.; Chesnel, F.; Guiguen, Y.; Bobe, J. Double maternal-effect: Duplicated nucleoplasmin 2 genes, npm2a and npm2b, with essential but distinct functions are shared by fish and tetrapods. BMC Evol. Biol. 2018, 18, 167. [Google Scholar] [CrossRef]
- Howley, C.; Ho, R.K. mRNA localization patterns in zebrafish oocytes. Mech. Dev. 2000, 92, 305–309. [Google Scholar] [CrossRef]
- Fuentes, R.; Fernández, J. Ooplasmic segregation in the zebrafish zygote and early embryo: Pattern of ooplasmic movements and transport pathways. Dev. Dyn. 2010, 239, 2172–2189. [Google Scholar] [CrossRef]
- Benítez-Santana, T.; Atalah, E.; Betancor, M.B.; Caballero, M.; Hernandez-Cruz, C.M.; Izquierdo, M. DHA but not EPA, enhances sound induced escape behavior and Mauthner cells activity in Sparus aurata. Physiol. Behav. 2014, 124, 65–71. [Google Scholar] [CrossRef]
- Karlstrom, R.O.; Kane, D.A. A flipbook of zebrafish embryogenesis. Development 1996, 123, 461. [Google Scholar] [PubMed]
- Poupard, G.; André, M.; Durliat, M.; Ballagny, C.; Boeuf, G.; Babin, P.J. Apolipoprotein E gene expression correlates with endogenous lipid nutrition and yolk syncytial layer lipoprotein synthesis during fish development. Cell Tissue Res. 2000, 300, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, M.D. Utilization of yolk fatty acids by goldfish embryos and larvae. Fish Physiol. Biochem. 1996, 15, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, M.; Fernández-Palacios, H.; Tacon, A. Effect of broodstock nutrition on reproductive performance of fish. Aquaculture 2001, 197, 25–42. [Google Scholar] [CrossRef]
- Izquierdo, M.S.; Robaina, L.; Juárez-Carrillo, E.; Oliva, V.; Hernández-Cruz, C.M.; Afonso, J.M. Regulation of growth, fatty acid composition and delta 6 desaturase expression by dietary lipids in gilthead seabream larvae (Sparus aurata). Fish Physiol. Biochem. 2008, 34, 117–127. [Google Scholar] [CrossRef]
- Li, S.; Mai, K.; Xu, W.; Yuan, Y.; Zhang, Y.; Ai, Q. Characterization, mRNA expression and regulation of Δ6 fatty acyl desaturase (FADS2) by dietary n−3 long chain polyunsaturated fatty acid (LC-PUFA) levels in grouper larvae (Epinephelus coioides). Aquaculture 2014, 434, 212–219. [Google Scholar] [CrossRef]
- Vagner, M.; Infante, J.Z.; Robin, J.; Ruyet, J.P.-L. Is it possible to influence European sea bass (Dicentrarchus labrax) juvenile metabolism by a nutritional conditioning during larval stage? Aquaculture 2007, 267, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Nara, T.Y.; He, W.S.; Tang, C.; Clarke, S.D.; Nakamura, M.T. The E-box like sterol regulatory element mediates the suppression of human Δ-6 desaturase gene by highly unsaturated fatty acids. Biochem. Biophys. Res. Commun. 2002, 296, 111–117. [Google Scholar] [CrossRef]
- Jaroszewska, M.; Dabrowski, K. Utilization of yolk: Transition from endogenous to exogenous nutrition in fish. In Larval Fish Nutrition; John Wiley & Sons, Inc.: Chichester, UK, 2011; pp. 183–218. [Google Scholar]
- Park, W.J.; Kothapalli, K.S.D.; Lawrence, P.; Tyburczy, C.; Brenna, J.T. An alternate pathway to long-chain polyunsaturates: The FADS2 gene product Δ8-desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 2009, 50, 1195–1202. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Palacios, H.; Izquierdo, M.S.; Robaina, L.; Valencia, A.; Salhi, M.; Vergara, J. Effect of n-3 HUFA level in broodstock diets on egg quality of gilthead sea bream (Sparus aurata L.). Aquaculture 1995, 132, 325–337. [Google Scholar] [CrossRef]
- Morais, S.; Mendes, A.C.; Castanheira, M.F.; Coutinho, J.; Bandarra, N.M.; Dias, J.; Conceicao, L.E.; Ferreira, P.P. New formulated diets for Solea senegalensis broodstock: Effects of parental nutrition on biosynthesis of long-chain polyunsaturated fatty acids and performance of early larval stages and juvenile fish. Aquaculture 2014, 432, 374–382. [Google Scholar] [CrossRef]





| Group | Gender | fads2 Expression (Copies/μL) |
|---|---|---|
| HRO (High fas2 + RO diet) | Female 1 | 14.92 |
| Male 1-1 | 7.06 | |
| Male 1-2 | 3.37 | |
| Female 2 | 12.72 | |
| Male 2-1 | 4.09 | |
| Male 2-2 | 3.33 | |
| LRO (Low fas2 + RO diet) | Female 3 | 1.41 |
| Male 3-1 | 1.53 | |
| Male 3-2 | 1.46 | |
| Female 4 | 0.49 | |
| Male 4-1 | 1.74 | |
| Male 4-2 | 1.66 | |
| LFO (low fas2 + FO diet) | Female 5 | 1.11 |
| Male 5-1 | 1.26 | |
| Male 5-2 | 1.27 |
| Fatty Acids | HRO | LRO | ||
|---|---|---|---|---|
| Mean | S.D. | Mean | S.D. | |
| 18:1n-9 | 27.06 | 1.03 | 19.69 | 7.04 |
| 18:2n-9 | 0.10 | 0.02 | 0.33 | 0.13 |
| 18.2n-6 | 10.81 | 0.24 | 8.63 | 1.76 |
| 18:3n-6 | 0.20 | 0.04 | 0.56 | 0.15 |
| 18:3n-3 | 2.92 | 0.12 | 2.20 | 0.53 |
| 18:4n-3 | 0.53 | 0.01 | 1.02 | 0.58 |
| 20:4n-6 | 0.76 | 0.03 | 0.78 | 0.01 |
| 20:5n-3 | 5.95 | 0.10 | 6.98 | 3.49 |
| 22:4n-6 | 0.08 | 0.02 | 0.63 * | 0.07 |
| 22:5n-6 | 0.25 | 0.04 | 0.60 * | 0.04 |
| 22:6n-3 | 20.45 * | 0.64 | 13.99 | 0.21 |
| SFA | 16.76 | 0.36 | 25.38 * | 0.31 |
| MUFA | 35.74 | 0.82 | 31.76 | 5.30 |
| PUFA | 47.49 | 1.20 | 42.87 | 4.97 |
| n-3 FA | 33.84 | 0.83 | 28.01 | 5.15 |
| n-6 FA | 12.67 | 0.38 | 12.06 | 1.42 |
| DHA/ARA | 26.91 * | 0.16 | 17.94 | 0.05 |
| DHA/EPA | 3.44 | 0.05 | 2.28 | 1.11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Ferosekhan, S.; Turkmen, S.; Afonso, J.M.; Zamorano, M.J.; Izquierdo, M. Influence of Parental Fatty Acid Desaturase 2 (fads2) Expression and Diet on Gilthead Seabream (Sparus aurata) Offspring fads2 Expression during Ontogenesis. Animals 2020, 10, 2191. https://doi.org/10.3390/ani10112191
Xu H, Ferosekhan S, Turkmen S, Afonso JM, Zamorano MJ, Izquierdo M. Influence of Parental Fatty Acid Desaturase 2 (fads2) Expression and Diet on Gilthead Seabream (Sparus aurata) Offspring fads2 Expression during Ontogenesis. Animals. 2020; 10(11):2191. https://doi.org/10.3390/ani10112191
Chicago/Turabian StyleXu, Hanlin, Shajahan Ferosekhan, Serhat Turkmen, Juan Manuel Afonso, María Jesús Zamorano, and Marisol Izquierdo. 2020. "Influence of Parental Fatty Acid Desaturase 2 (fads2) Expression and Diet on Gilthead Seabream (Sparus aurata) Offspring fads2 Expression during Ontogenesis" Animals 10, no. 11: 2191. https://doi.org/10.3390/ani10112191
APA StyleXu, H., Ferosekhan, S., Turkmen, S., Afonso, J. M., Zamorano, M. J., & Izquierdo, M. (2020). Influence of Parental Fatty Acid Desaturase 2 (fads2) Expression and Diet on Gilthead Seabream (Sparus aurata) Offspring fads2 Expression during Ontogenesis. Animals, 10(11), 2191. https://doi.org/10.3390/ani10112191








