Genomic Analysis of Milk Protein Fractions in Brown Swiss Cattle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Phenotypic and Genotypic Information
2.3. Statistical Analysis
2.4. Genetic Parameters
3. Results
3.1. Descriptive Statistics
3.2. Genetic Parameters
3.3. Genetic Correlations
3.4. Phenotypic Correlations
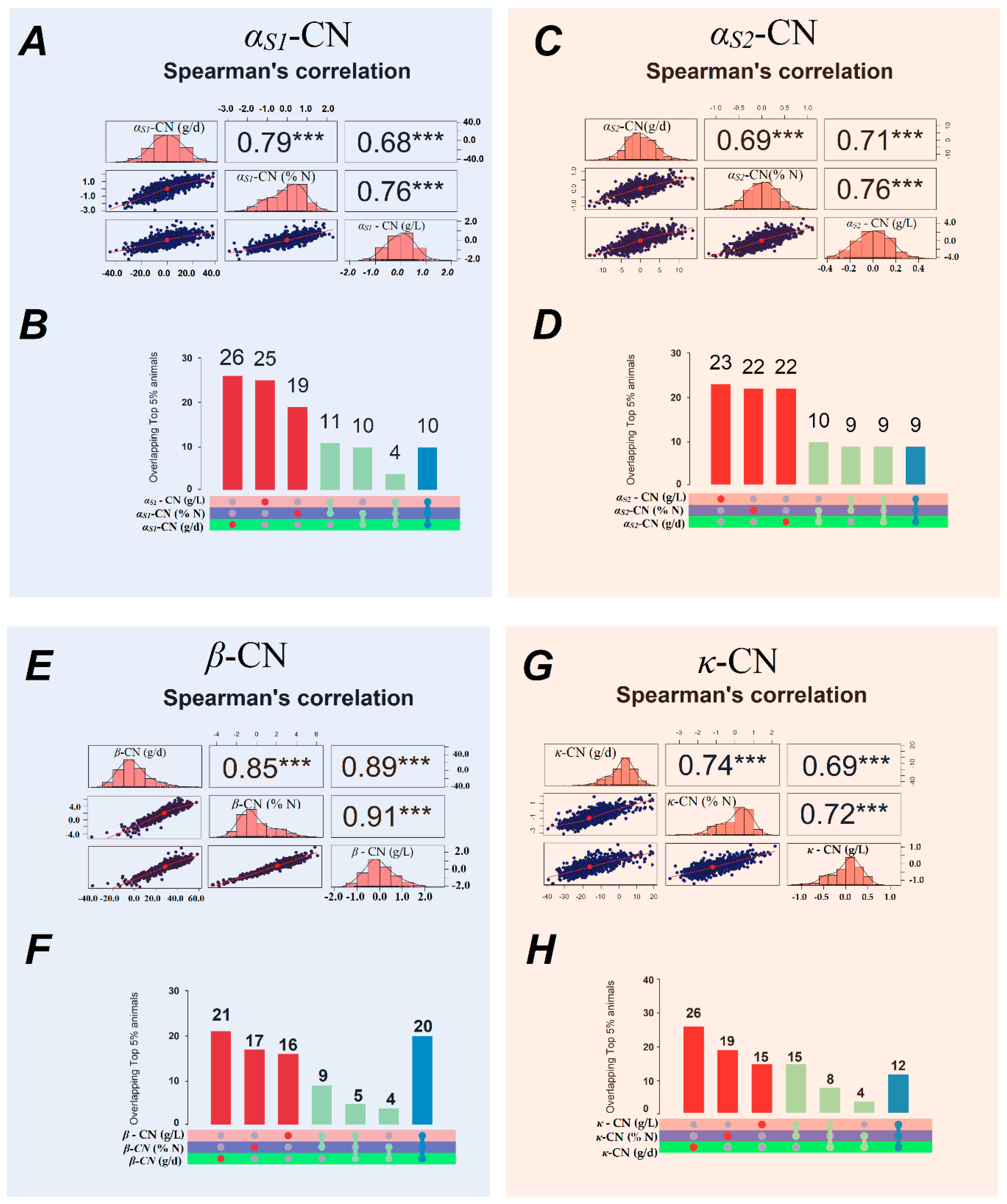
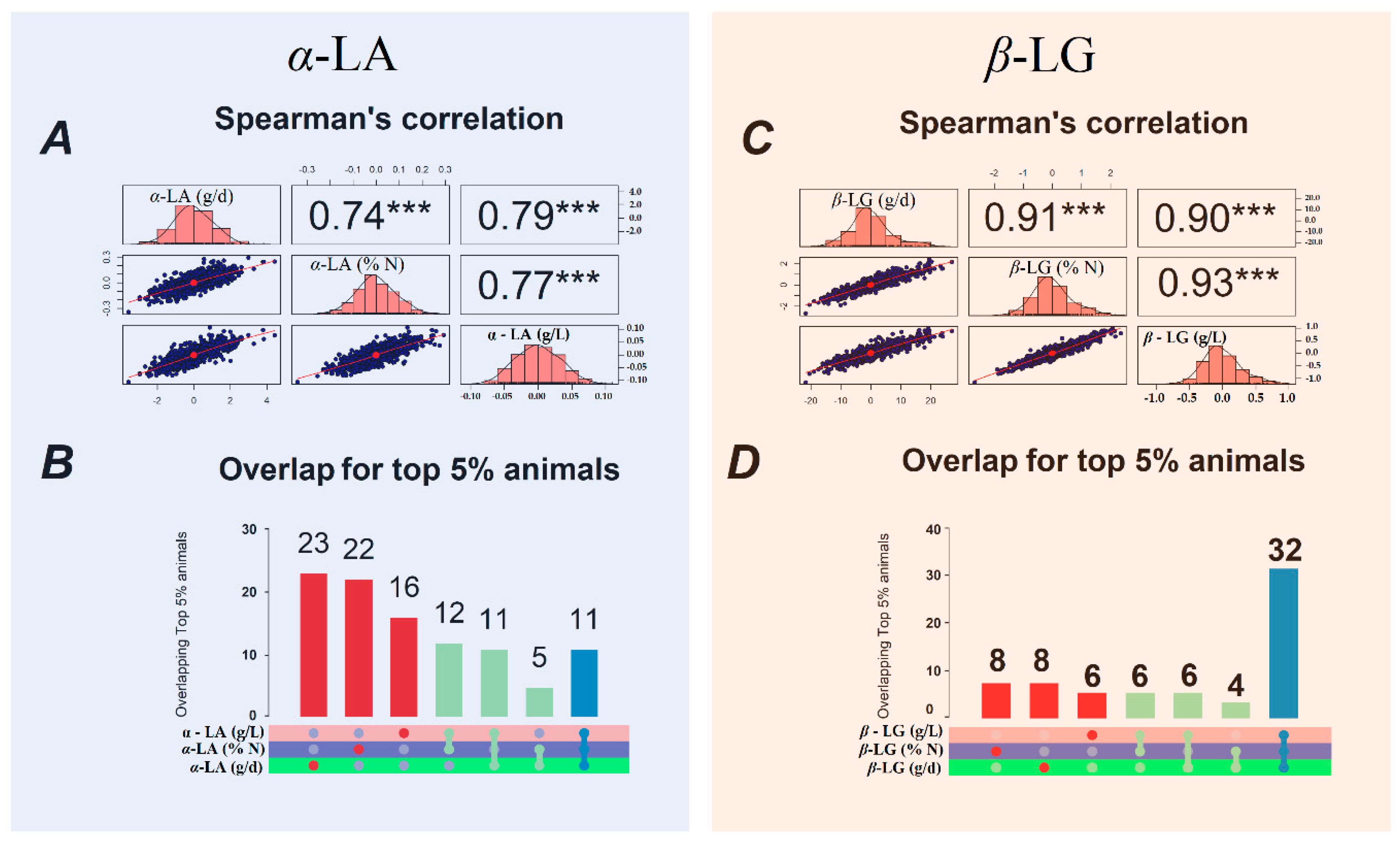
3.5. Spearman’s Correlations
4. Discussion
4.1. Genetic Parameters
4.2. Genetic and Phenotypic Correlations
4.3. Spearman’s Correlations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wedholm, A.; Larsen, L.B.; Lindmark-Månsson, H.; Karlsson, A.H.; Andrén, A. Effect of Protein Composition on the Cheese-Making Properties of Milk from Individual Dairy Cows. J. Dairy Sci. 2006, 89, 3296–3305. [Google Scholar] [CrossRef]
- Holland, B.; Rahimi Yazdi, S.; Ion Titapiccolo, G.; Corredig, M. Short communication: Separation and quantification of caseins and casein macropeptide using ion-exchange chromatography. J. Dairy Sci. 2010, 93, 893–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonfatti, V.; Cecchinato, A.; Gallo, L.; Blasco, A.; Carnier, P. Genetic analysis of detailed milk protein composition and coagulation properties in Simmental cattle. J. Dairy Sci. 2011, 94, 5183–5193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferragina, A.; de los Campos, G.; Vazquez, A.I.; Cecchinato, A.; Bittante, G. Bayesian regression models outperform partial least squares methods for predicting milk components and technological properties using infrared spectral data. J. Dairy Sci. 2015, 98, 8133–8151. [Google Scholar] [CrossRef] [Green Version]
- Schopen, G.C.B.; Heck, J.M.L.; Bovenhuis, H.; Visker, M.H.P.W.; van Valenberg, H.J.F.; van Arendonk, J.A.M. Genetic parameters for major milk proteins in Dutch Holstein-Friesians. J. Dairy Sci. 2009, 92, 1182–1191. [Google Scholar] [CrossRef]
- Cecchinato, A.; Ribeca, C.; Maurmayr, A.; Penasa, M.; De Marchi, M.; Macciotta, N.P.P.; Mele, M.; Secchiari, P.; Pagnacco, G.; Bittante, G. Effects of β-lactoglobulin, stearoyl-coenzyme A desaturase 1, and sterol regulatory element binding protein gene allelic variants on milk production, composition, acidity, and coagulation properties of Brown Swiss cows. J. Dairy Sci. 2012, 95, 450–454. [Google Scholar] [CrossRef]
- Dadousis, C.; Cipolat-Gotet, C.; Bittante, G.; Cecchinato, A. Inferring genetic parameters on latent variables underlying milk yield and quality, protein composition, curd firmness and cheese-making traits in dairy cattle. Animal 2018, 12, 224–231. [Google Scholar] [CrossRef]
- Gebreyesus, G.; Lund, M.S.; Janss, L.; Poulsen, N.A.; Larsen, L.B.; Bovenhuis, H.; Buitenhuis, A.J. Multi-trait estimation of genetic parameters for milk protein composition in the Danish Holstein. J. Dairy Sci. 2016, 99, 2863–2866. [Google Scholar] [CrossRef] [Green Version]
- Pegolo, S.; Mach, N.; Ramayo-Caldas, Y.; Schiavon, S.; Bittante, G.; Cecchinato, A. Integration of GWAS, pathway and network analyses reveals novel mechanistic insights into the synthesis of milk proteins in dairy cows. Sci. Rep. 2018, 8, 566. [Google Scholar] [CrossRef]
- Sanchez, M.P.; Ferrand, M.; Gelé, M.; Pourchet, D.; Miranda, G.; Martin, P.; Brochard, M.; Boichard, D. Genetic parameters for milk protein composition predicted using mid-infrared spectroscopy in the French Montbéliarde, Normande, and Holstein dairy cattle breeds. J. Dairy Sci. 2017, 100, 6371–6375. [Google Scholar] [CrossRef]
- Bittante, G.; Cipolat-Gotet, C.; Malchiodi, F.; Sturaro, E.; Tagliapietra, F.; Schiavon, S.; Cecchinato, A. Effect of dairy farming system, herd, season, parity, and days in milk on modeling of the coagulation, curd firming, and syneresis of bovine milk. J. Dairy Sci. 2015, 98, 2759–2774. [Google Scholar] [CrossRef] [PubMed]
- Cecchinato, A.; Albera, A.; Cipolat-Gotet, C.; Ferragina, A.; Bittante, G. Genetic parameters of cheese yield and curd nutrient recovery or whey loss traits predicted using Fourier-transform infrared spectroscopy of samples collected during milk recording on Holstein, Brown Swiss, and Simmental dairy cows. J. Dairy Sci. 2015, 98, 4914–4927. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.K.A.K.A.; Shook, G.E.E. An Optimum Transformation for Somatic Cell Concentration in Milk. J. Dairy Sci. 1980, 63, 487–490. [Google Scholar] [CrossRef]
- Bonfatti, V.; Grigoletto, L.; Cecchinato, A.; Gallo, L.; Carnier, P. Validation of a new reversed-phase high-performance liquid chromatography method for separation and quantification of bovine milk protein genetic variants. J. Chromatogr. A 2008, 1195, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Perdry, H.; Dandine-Roulland, L. Gaston—Genetic Data Handling (QC, GRM, LD, PCA) & Linear Mixed Models. R Packag. 2018, 83, 1–29. [Google Scholar]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [Green Version]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs; University of Georgia: Athens, GA, USA, 2015. [Google Scholar]
- Bonfatti, V.; Di Martino, G.; Cecchinato, A.; Vicario, D.; Carnier, P. Effects of β-κ-casein (CSN2-CSN3) haplotypes and β-lactoglobulin (BLG) genotypes on milk production traits and detailed protein composition of individual milk of Simmental cows. J. Dairy Sci. 2010, 93, 3797–3808. [Google Scholar] [CrossRef]
- Cipolat-Gotet, C.; Cecchinato, A.; Malacarne, M.; Bittante, G.; Summer, A. Variations in milk protein fractions affect the efficiency of the cheese-making process. J. Dairy Sci. 2018, 101, 8788–8804. [Google Scholar] [CrossRef] [Green Version]
- Amalfitano, N.; Cipolat-Gotet, C.; Cecchinato, A.; Malacarne, M.; Summer, A.; Bittante, G. Milk protein fractions strongly affect the patterns of coagulation, curd firming, and syneresis. J. Dairy Sci. 2019, 102, 2903–2917. [Google Scholar] [CrossRef] [Green Version]
- Heck, J.M.L.L.; Schennink, A.; Van Valenberg, H.J.F.F.; Bovenhuis, H.; Visker, M.H.P.W.P.W.; Van Arendonk, J.A.M.M.; Van Hooijdonk, A.C.M.M. Effects of milk protein variants on the protein composition of bovine milk. J. Dairy Sci. 2009, 92, 1192–1202. [Google Scholar] [CrossRef]
- Gustavsson, F.; Buitenhuis, A.; Johansson, M.; Bertelsen, H.; Glantz, M.; Poulsen, N.; Lindmark Månsson, H.; Stålhammar, H.; Larsen, L.; Bendixen, C.; et al. Effects of breed and casein genetic variants on protein profile in milk from Swedish Red, Danish Holstein, and Danish Jersey cows. J. Dairy Sci. 2014, 97, 3866–3877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobe, G.; Beitz, D.C.; Freeman, A.E.; Lindberg, G.L. Effect of Milk Protein Genotypes on Milk Protein Composition and Its Genetic Parameter Estimates. J. Dairy Sci. 1999, 82, 2797–2804. [Google Scholar] [CrossRef]
- Buitenhuis, B.; Poulsen, N.A.; Gebreyesus, G.; Larsen, L.B. Estimation of genetic parameters and detection of chromosomal regions affecting the major milk proteins and their post translational modifications in Danish Holstein and Danish Jersey cattle. BMC Genet. 2016, 17, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheverud, J.M. Quantitative genetics and developmental constraints on evolution by selection. J. Theor. Biol. 1984, 110, 155–171. [Google Scholar] [CrossRef]
- Sodini, S.M.; Kemper, K.E.; Wray, N.R.; Trzaskowski, M. Comparison of genotypic and phenotypic correlations: Cheverud’s conjecture in humans. Genetics 2018, 209, 941–948. [Google Scholar] [CrossRef]
- Bevilacqua, C.; Helbling, J.C.; Miranda, G.; Martin, P. Translational efficiency of casein transcripts in the mammary tissue of lactating ruminants. Reprod. Nutr. Dev. 2006, 46, 567–578. [Google Scholar] [CrossRef] [Green Version]
- Schopen, G.C.B.; Visker, M.H.P.W.; Koks, P.D.; Mullaart, E.; van Arendonk, J.A.M.; Bovenhuis, H. Whole-genome association study for milk protein composition in dairy cattle. J. Dairy Sci. 2011, 94, 3148–3158. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Peñagaricano, F.; Ahmad, K.R.; Lucey, J.A.; Weigel, K.A.; Khatib, H. Association between milk protein gene variants and protein composition traits in dairy cattle. J. Dairy Sci. 2012, 95, 440–449. [Google Scholar] [CrossRef]
- van Binsbergen, R.; Veerkamp, R.F.; Calus, M.P.L. Makeup of the genetic correlation between milk production traits using genome-wide single nucleotide polymorphism information. J. Dairy Sci. 2012, 95, 2132–2143. [Google Scholar] [CrossRef]
- Jensen, H.B.; Poulsen, N.A.; Andersen, K.K.; Hammershøj, M.; Poulsen, H.D.; Larsen, L.B. Distinct composition of bovine milk from Jersey and Holstein-Friesian cows with good, poor, or noncoagulation properties as reflected in protein genetic variants and isoforms. J. Dairy Sci. 2012, 95, 6905–6917. [Google Scholar] [CrossRef]
- Elofsson, U.M.; Paulsson, M.A.; Sellers, P.; Arnebrant, T. Adsorption during Heat Treatment Related to the Thermal Unfolding/Aggregation of β-Lactoglobulins A and B. J. Colloid Interface Sci. 1996, 183, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Molina, J.M.; Kramm, J.; Brito, B.C.; Carrillo, B.; Pinto, M.; Ferrando, A. Protein composition of milk from Holstein–Friesian dairy cows and its relationship with the genetic variants A and B of κ-casein and β-lactoglobulin (Part I). Int. J. Dairy Technol. 2006, 59, 183–187. [Google Scholar] [CrossRef]
| Trait1 | N | Mean | Min | Max | SD | V.C. |
|---|---|---|---|---|---|---|
| Milk yield (kg/d) | 981 | 24.87 | 5.91 | 45.33 | 7.28 | 29.27 |
| Protein fraction contents (g/L) | ||||||
| Major protein fractions | 983 | 32.07 | 22.90 | 42.21 | 3.49 | 10.88 |
| Major caseins | 983 | 28.03 | 19.94 | 36.90 | 2.97 | 10.59 |
| αS1-CN | 987 | 9.41 | 5.80 | 12.99 | 1.20 | 12.75 |
| αS2-CN | 986 | 3.37 | 1.98 | 5.11 | 0.57 | 16.91 |
| β-CN | 985 | 11.79 | 8.11 | 16.02 | 1.39 | 11.79 |
| κ-CN | 984 | 3.47 | 1.36 | 5.53 | 0.68 | 19.59 |
| Major whey proteins | 981 | 4.05 | 1.76 | 6.26 | 0.77 | 19.01 |
| α-LA | 982 | 0.87 | 0.38 | 1.42 | 0.18 | 20.69 |
| β-LG | 988 | 3.19 | 1.15 | 5.27 | 0.70 | 21.94 |
| Crude protein (g/L) | 985 | 36.60 | 25.80 | 48.40 | 4.10 | 10.79 |
| Protein fraction proportions (%N) | ||||||
| Major protein fractions | 980 | 87.67 | 81.77 | 93.55 | 2.25 | 2.57 |
| Major caseins | 981 | 76.62 | 73.27 | 79.83 | 1.24 | 1.62 |
| αS1-CN | 985 | 25.71 | 20.55 | 30.91 | 1.71 | 6.65 |
| αS2-CN | 979 | 9.17 | 6.26 | 12.58 | 1.05 | 11.45 |
| β-CN | 976 | 32.22 | 25.97 | 39.41 | 2.38 | 7.39 |
| κ-CN | 985 | 9.52 | 5.02 | 13.52 | 1.39 | 14.60 |
| Major whey proteins | 979 | 11.05 | 6.69 | 15.46 | 1.52 | 13.76 |
| α-LA | 983 | 2.39 | 1.03 | 3.81 | 0.48 | 20.08 |
| β-LG | 978 | 8.66 | 4.08 | 13.37 | 1.47 | 16.97 |
| Protein fraction daily yields (g/d) | ||||||
| Major protein fractions | 986 | 787.44 | 218.68 | 1370.19 | 225.67 | 28.66 |
| Major caseins | 984 | 687.76 | 191.33 | 847.99 | 95.75 | 13.92 |
| αS1-CN | 983 | 230.58 | 46.07 | 431.04 | 72.18 | 31.30 |
| αS2-CN | 987 | 83.07 | 15.73 | 163.49 | 26.95 | 32.44 |
| β-CN | 984 | 289.48 | 75.78 | 543.91 | 81.02 | 27.99 |
| κ-CN | 984 | 84.63 | 11.42 | 169.78 | 27.76 | 32.80 |
| Major whey proteins | 983 | 99.68 | 21.36 | 190.73 | 33.11 | 33.21 |
| α-LA | 983 | 21.64 | 4.01 | 47.97 | 8.71 | 40.25 |
| β-LG | 985 | 78.04 | 15.11 | 159.48 | 27.02 | 34.62 |
| Trait | |||
|---|---|---|---|
| Milk yield (kg/d) | 2.79 (0.892) | 22.09 (1.256) | 0.12 (0.033) |
| Protein fraction contents (g/L) | |||
| Major protein fractions | 2.41 (0.020) | 6.07 (0.094) | 0.39 (0.007) |
| Major caseins | 1.68 (0.045) | 4.41 (0.088) | 0.38 (0.009) |
| αS1-CN | 0.51 (0.002) | 0.92 (0.007) | 0.55 (0.001) |
| αS2-CN | 0.05 (0.001) | 0.18 (0.009) | 0.27 (0.002) |
| β-CN | 0.64 (0.002) | 1.08 (0.005) | 0.59 (0.004) |
| κ-CN | 0.18 (0.007) | 0.33 (0.015) | 0.55 (0.005) |
| Major whey proteins | 0.12 (0.003) | 0.29 (0.012) | 0.43 (0.005) |
| α-LA | 0.003 (0.000)* | 0.014 (0.000)* | 0.21 (0.007) |
| β-LG | 0.12 (0.001) | 0.24 (0.008) | 0.53 (0.006) |
| Crude protein (g/L) | 0.36 (0.007) | 0.82 (0.06) | 0.44 (0.06) |
| Protein fraction proportions (%N) | |||
| Major protein fractions | 0.84 (0.013) | 2.07 (0.023) | 0.40 (0.009) |
| Major caseins | 0.37 (0.054) | 1.01 (0.068) | 0.37 (0.009) |
| αS1-CN | 1.20 (0.003) | 2.06 (0.066) | 0.58 (0.009) |
| αS2-CN | 0.23 (0.002) | 0.80 (0.018) | 0.29 (0.008) |
| β-CN | 3.67 (0.004) | 4.69 (0.092) | 0.78 (0.003) |
| κ-CN | 1.10 (0.009) | 1.83 (0.081) | 0.60 (0.006) |
| Major whey proteins | 0.56 (0.010) | 1.29 (0.082) | 0.43 (0.011) |
| α-LA | 0.02 (0.003) | 0.11 (0.022) | 0.23 (0.008) |
| β-LG | 0.63 (0.003) | 1.20 (0.045) | 0.53 (0.005) |
| Protein fraction daily yields (g/d) | |||
| Major protein fractions | 283.60 (1.206) | 2015.45 (1.348) | 0.15 (0.004) |
| Major caseins | 207.47 (0.985) | 1320.91 (1.055) | 0.14 (0.007) |
| αS1-CN | 285.35 (2.578) | 1284.45 (17.086) | 0.25 (0.002) |
| αS2-CN | 52.60 (0.231) | 258.12 (6.345) | 0.20 (0.009) |
| β-CN | 521.10 (2.739) | 2334.14 (16.226) | 0.22 (0.002) |
| κ-CN | 105.65 (0.380) | 357.63 (3.469) | 0.29 (0.001) |
| Major whey proteins | 90.81 (0.029) | 340.66 (0.832) | 0.21 (0.007) |
| α-LA | 5.45 (0.020) | 23.47 (0.679) | 0.23 (0.007) |
| β-LG | 97.09 (0.353) | 256.29 (4.904) | 0.38 (0.003) |
| Protein Fraction Contents (g/L) | |||||||
| Title | MY | αS1-CN | αS2-CN | β-CN | κ-CN | α-LA | β-LG |
| MY | - | 0.27 (0.007) | −0.31 (0.004) | −0.42 (0.006) | −0.27 (0.008) | −0.20 (0.005) | −0.15 (0.005) |
| αS1-CN | 0.02 (0.004) | - | 0.74 (0.002) | −0.30 (0.002) | 0.55 (0.001) | 0.44 (0.001) | 0.56 (0.001) |
| αS2-CN | −0.29 (0.001) | 0.55 (0.006) | - | −0.23 (0.001) | 0.34 (0.002) | 0.71 (0.001) | 0.34 (0.004) |
| β-CN | −0.63 (0.004) | −0.36 (0.002) | −0.25 (0.002) | - | −0.48 (0.004) | −0.12 (0.002) | 0.17 (0.003) |
| κ-CN | −0.14 (0.002) | 0.44 (0.004) | 0.26 (0.005) | −0.03 (0.002) | - | 0.21 (0.003) | 0.33 (0.006) |
| α-LA | −0.04 (0.001) | 0.33 (0.002) | 0.42 (0.003) | 0.19 (0.002) | −0.11 (0.002) | - | 0.42 (0.001) |
| β-LG | −0.26 (0.002) | 0.55 (0.002) | 0.45 (0.004) | 0.30 (0.003) | 0.32 (0.004) | 0.38 (0.002) | - |
| Title | Protein Fraction Proportions (%N) | ||||||
| CP | αS1-CN | αS2-CN | β-CN | κ-CN | α-LA | β-LG | |
| CP | - | −0.41 (0.002) | −0.15 (0.003) | −0.25 (0.002) | 0.17 (0.003) | −0.50 (0.004) | 0.18 (0.002) |
| αS1-CN | 0.16 (0.021) | - | 0.59 (0.001) | −0.70 (0.005) | 0.56 (0.001) | 0.38 (0.001) | 0.25 (0.003) |
| αS2-CN | 0.07 (0.005) | 0.09 (0.005) | - | −0.43 (0.002) | 0.36 (0.002) | 0.67 (0.002) | 0.23 (0.001) |
| β-CN | −0.20 (0.003) | −0.58 (0.005) | −0.35 (0.008) | - | −0.78 (0.001) | 0.20 (0.002) | −0.23 (0.002) |
| κ-CN | 0.09 (0.005) | 0.08 (0.003) | 0.11 (0.007) | −0.57 (0.004) | - | 0.14 (0.001) | −0.35 (0.002) |
| α-LA | −0.25 (0.001) | 0.05 (0.002) | 0.25 (0.002) | 0.04 (0.009) | −0.13 (0.005) | - | 0.40 (0.002) |
| β-LG | 0.09 (0.002) | 0.18 (0.002) | 0.16 (0.003) | −0.18 (0.006) | −0.02 (0.005) | 0.24 (0.003) | - |
| Protein Fraction Daily Yields (g/d) | |||||||
| αS1-CN | αS2-CN | β-CN | κ-CN | α-LA | β-LG | ||
| αS1-CN | - | - | 0.85 (0.006) | 0.42 (0.001) | 0.51 (0.004) | 0.63 (0.020) | 0.61 (0.008) |
| αS2-CN | - | 0.84 (0.008) | - | 0.38 (0.001) | 0.39 (0.003) | 0.70 (0.012) | 0.47 (0.005) |
| β-CN | - | 0.84 (0.002) | 0.77 (0.003) | - | −0.30 (0.003) | 0.45 (0.018) | 0.30 (0.005) |
| κ-CN | - | 0.75 (0.004) | 0.63 (0.008) | −0.61 (0.003) | - | 0.07 (0.009) | 0.37 (0.001) |
| α-LA | - | 0.77 (0.026) | 0.76 (0.009) | 0.74 (0.009) | 0.57 (0.010) | - | 0.58 (0.030) |
| β-LG | - | 0.79 (0.002) | 0.73 (0.007) | 0.72 (0.002) | 0.64 (0.005) | 0.71 (0.004) | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macedo Mota, L.F.; Pegolo, S.; Bisutti, V.; Bittante, G.; Cecchinato, A. Genomic Analysis of Milk Protein Fractions in Brown Swiss Cattle. Animals 2020, 10, 336. https://doi.org/10.3390/ani10020336
Macedo Mota LF, Pegolo S, Bisutti V, Bittante G, Cecchinato A. Genomic Analysis of Milk Protein Fractions in Brown Swiss Cattle. Animals. 2020; 10(2):336. https://doi.org/10.3390/ani10020336
Chicago/Turabian StyleMacedo Mota, Lucio Flavio, Sara Pegolo, Vittoria Bisutti, Giovanni Bittante, and Alessio Cecchinato. 2020. "Genomic Analysis of Milk Protein Fractions in Brown Swiss Cattle" Animals 10, no. 2: 336. https://doi.org/10.3390/ani10020336






