Repeatability and Reproducibility of Measures of Bovine Methane Emissions Recorded using a Laser Detector
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Measurements of CH4
2.2. Data Editing
2.3. Repeatability and Reproducibility of Phenotypes
3. Results and Discussion
3.1. Distribution and Descriptive Statistics
3.2. Mean and Aggregate of CH4 Emissions
3.3. Peaks of CH4 Emissions
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- IPCC. Climate Change 2014: Synthesis Report; Intergovernmental Panel on Climate Change (IPCC): Geneva, Switzerland, 2014; Available online: https://archive.ipcc.ch/report/ar5/syr/ (accessed on 26 March 2020).
- Steinfeld, H.; Gerber, P.; Wassenaar, T.; Castel, V.; Rosales, M.; De Haan, C. Livestock’s long shadow; FAO: Rome, Italy, 2006; Available online: http://www.fao.org/3/a0701e/a0701e00.htm (accessed on 15 February 2020).
- Cassandro, M.; Mele, M.; Stefanon, B. Genetic aspects of enteric methane emission in ruminants livestock. Ital. J. Anim. Sci. 2013, 12, 450–458. [Google Scholar] [CrossRef]
- Johnson, K.A. Methane emissions from cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Hristov, A.; Kebreab, E.; Niu, M.; Oh, J.; Bannink, A.; Bayat, A.R.; Boland, T.; Brito, A.; Casper, D.; Crompton, L.; et al. Symposium review: Uncertainties in enteric methane inventories, measurement techniques, and prediction models. J. Dairy Sci. 2018, 101, 6655–6674. [Google Scholar] [CrossRef] [PubMed]
- Dehareng, F.; Delfosse, C.; Froidmont, E.; Soyeurt, H.; Martin, C.; Gengler, N.; Vanlierde, A.; Dardenne, P. Potential use of milk mid-infrared spectra to predict individual methane emission of dairy cows. Anim. 2012, 6, 1694–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCartney, C.; Bull, I.; Yan, T.; Dewhurst, R. Assessment of archaeol as a molecular proxy for methane production in cattle. J. Dairy Sci. 2013, 96, 1211–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammond, K.; Crompton, L.; Bannink, A.; Dijkstra, J.; Yáñez-Ruiz, D.; O’Kiely, P.; Kebreab, E.; Eugène, M.; Yu, Z.; Shingfield, K.; et al. Review of current in vivo measurement techniques for quantifying enteric methane emission from ruminants. Anim. Feed. Sci. Technol. 2016, 219, 13–30. [Google Scholar] [CrossRef] [Green Version]
- Chagunda, M.G.G.; Yan, T. Do methane measurements from a laser detector and an indirect open-circuit respiration calorimetric chamber agree sufficiently closely? Anim. Feed. Sci. Technol. 2011, 165, 8–14. [Google Scholar] [CrossRef]
- Sorg, D.; Mühlbach, S.; Rosner, F.; Kuhla, B.; Derno, M.; Meese, S.; Schwarm, A.; Kreuzer, M.; Swalve, H. The agreement between two next-generation laser methane detectors and respiration chamber facilities in recording methane concentrations in the spent air produced by dairy cows. Comput. Electron. Agric. 2017, 143, 262–272. [Google Scholar] [CrossRef]
- Sorg, D.; Difford, G.F.; Mühlbach, S.; Kuhla, B.; Swalve, H.; Lassen, J.; Strabel, T.; Pszczola, M. Comparison of a laser methane detector with the GreenFeed and two breath analysers for on-farm measurements of methane emissions from dairy cows. Comput. Electron. Agric. 2018, 153, 285–294. [Google Scholar] [CrossRef]
- Chagunda, M.G.G.; Ross, D.; Roberts, D. On the use of a laser methane detector in dairy cows. Comput. Electron. Agric. 2009, 68, 157–160. [Google Scholar] [CrossRef]
- Franzoi, M.; Niero, G.; Penasa, M.; Cassandro, M.; De Marchi, M. Technical note: Development and validation of a new method for the quantification of soluble and micellar calcium, magnesium, and potassium in milk. J. Dairy Sci. 2018, 101, 1883–1888. [Google Scholar] [CrossRef] [PubMed]
- Niero, G.; Penasa, M.; Berard, J.; Kreuzer, M.; Cassandro, M.; De Marchi, M. Technical note: Development and validation of an HPLC method for the quantification of tocopherols in different types of commercial cow milk. J. Dairy Sci. 2018, 101, 6866–6871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Shook, G. An Optimum Transformation for Somatic Cell Concentration in Milk. J. Dairy Sci. 1980, 63, 487–490. [Google Scholar] [CrossRef]
- Benedet, A.; Franzoi, M.; Penasa, M.; Pellattiero, E.; De Marchi, M. Prediction of blood metabolites from milk mid-infrared spectra in early-lactation cows. J. Dairy Sci. 2019, 102, 11298–11307. [Google Scholar] [CrossRef] [PubMed]
- Niero, G.; Franzoi, M.; Vigolo, V.; Penasa, M.; Cassandro, M.; Boselli, C.; Giangolini, G.; De Marchi, M. Validation of a gold standard method for iodine quantification in raw and processed milk, and its variation in different dairy species. J. Dairy Sci. 2019, 102, 4808–4815. [Google Scholar] [CrossRef] [PubMed]
- Bobbo, T.; Penasa, M.; Finocchiaro, R.; Visentin, G.; Cassandro, M. Alternative somatic cell count traits exploitable in genetic selection for mastitis resistance in Italian Holsteins. J. Dairy Sci. 2018, 101, 10001–10010. [Google Scholar] [CrossRef] [PubMed]
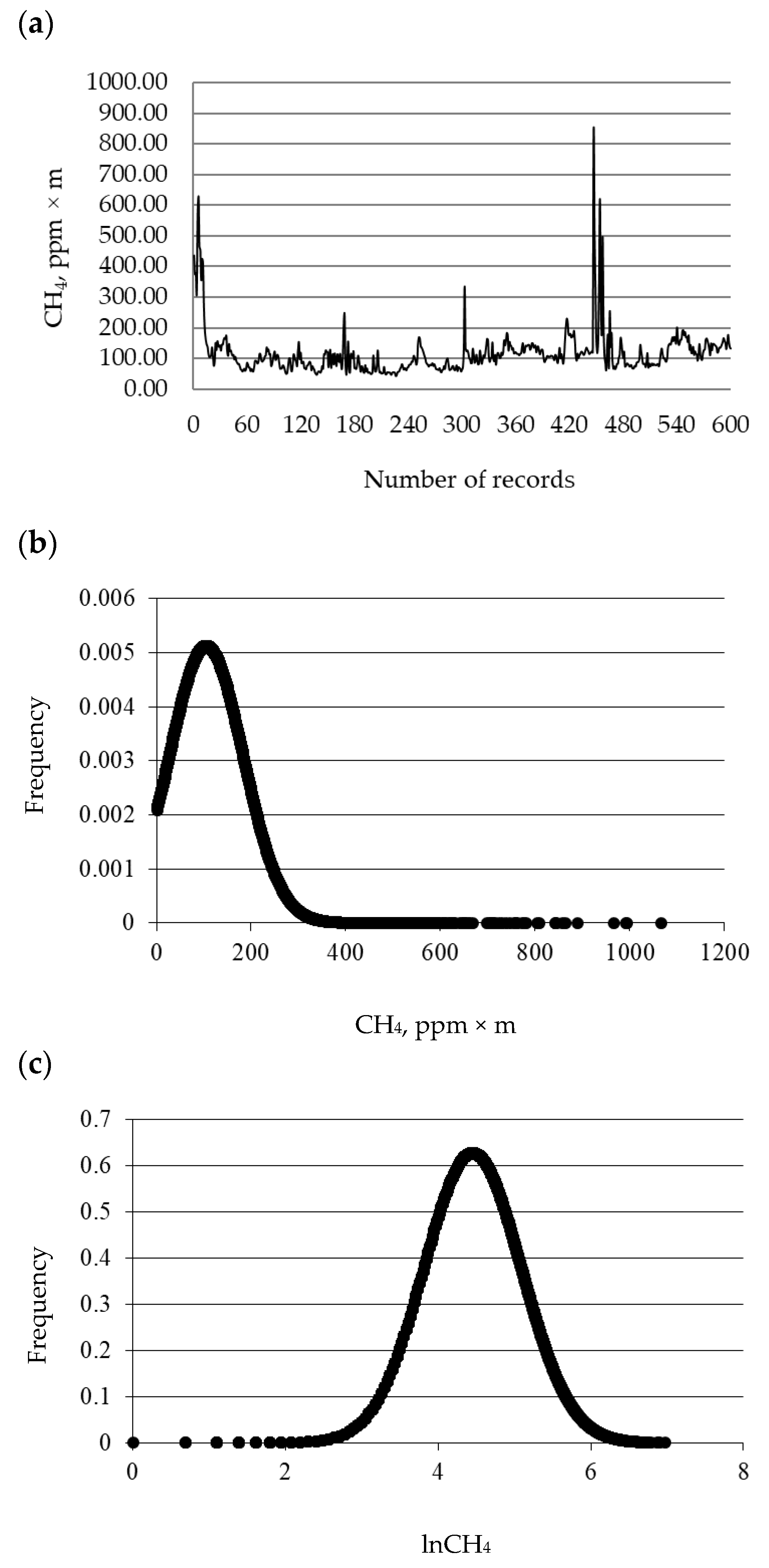
| Item | n | Mean | Mode | Median | Skewness | Kurtosis | Minimum | Maximum | SD 1 |
|---|---|---|---|---|---|---|---|---|---|
| CH4, ppm × m | |||||||||
| Pre-editing | 27,000 | 105.48 | 64 | 88 | 2.92 | 14.73 | 1.00 | 1067.00 | 77.92 |
| Post-editing | 26,449 | 98.26 | 64 | 86 | 1.36 | 2.18 | 1.00 | 339.00 | 58.02 |
| lnCH4 | |||||||||
| Pre-editing | 27,000 | 4.45 | 4.16 | 4.48 | −0.08 | 0.98 | 0.00 | 6.97 | 0.64 |
| Post-editing | 26,856 | 4.46 | 4.16 | 4.48 | 0.06 | 0.08 | 2.57 | 6.37 | 0.61 |
| Item | Day 1 | Day 2 | Day 3 | Overall | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Records | Mean | RSDr | Records | Mean | RSDr | Records | Mean | RSDr | Records | Mean | RSDR | |
| CH4, ppm × m | ||||||||||||
| Heifer 1 | 2941 | 84.61 | 67.03 | 2897 | 125.34 | 45.48 | 2913 | 123.76 | 41.57 | 8751 | 111.12 | 52.43 |
| Heifer 2 | 2943 | 95.76 | 57.31 | 2925 | 97.90 | 57.23 | 2945 | 51.34 | 74.48 | 8813 | 104.07 | 56.03 |
| Heifer 3 | 2988 | 51.33 | 74.48 | 2946 | 110.40 | 50.23 | 2951 | 78.14 | 67.84 | 8885 | 79.81 | 55.01 |
| Overall | 8872 | 77.10 | 70.03 | 8768 | 111.16 | 51.51 | 8809 | 106.72 | 53.17 | 26,449 | 98.26 | 59.05 |
| lnCH4 | ||||||||||||
| Heifer 1 | 2985 | 4.28 | 14.85 | 2992 | 4.78 | 9.68 | 2985 | 4.77 | 8.93 | 8962 | 4.61 | 12.29 |
| Heifer 2 | 2982 | 4.46 | 12.25 | 2980 | 4.48 | 12.58 | 2997 | 4.70 | 10.40 | 8959 | 4.55 | 11.98 |
| Heifer 3 | 2962 | 3.79 | 14.68 | 2995 | 4.63 | 10.18 | 2978 | 4.22 | 14.41 | 8935 | 4.21 | 15.35 |
| Overall | 8929 | 4.18 | 15.48 | 8967 | 4.63 | 11.15 | 8960 | 4.56 | 12.47 | 26,856 | 4.46 | 13.76 |
| Animals | Day 1 | Day 2 | Day 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Records | Aggregate | Slope | R2 | Records | Aggregate | Slope | R2 | Records | Aggregate | Slope | R2 | |
| CH4, ppm × m | ||||||||||||
| Heifer 1 | 2941 | 248,837 | 83.16 | 0.991 | 2897 | 363,110 | 129.77 | 0.997 | 2913 | 360,499 | 127.42 | 0.997 |
| Heifer 2 | 2943 | 281,808 | 101.84 | 0.997 | 2925 | 286,357 | 99.63 | 0.989 | 2945 | 349,011 | 125.67 | 0.996 |
| Heifer 3 | 2988 | 153,393 | 52.06 | 0.997 | 2946 | 325,148 | 111.60 | 0.997 | 2951 | 230,607 | 80.08 | 0.996 |
| lnCH4 | ||||||||||||
| Heifer 1 | 2985 | 12,787 | 4.26 | 0.999 | 2992 | 14,313 | 4.82 | 0.999 | 2985 | 14,250 | 4.80 | 0.999 |
| Heifer 2 | 2982 | 13,289 | 4.52 | 0.999 | 2980 | 13,354 | 4.49 | 0.999 | 2997 | 14,082 | 4.77 | 0.999 |
| Heifer 3 | 2962 | 11,212 | 3.79 | 0.999 | 2995 | 13,858 | 4.64 | 0.999 | 2978 | 12,562 | 4.26 | 0.999 |
| Item | Day 1 | Day 2 | Day 3 | Overall | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Records | Mean | RSDr | Records | Mean | RSDr | Records | Mean | RSDr | Records | Mean | RSDR | |
| CH4, ppm × m | ||||||||||||
| Heifer 1 | 223 | 235.89 | 18.79 | 468 | 231.16 | 19.88 | 427 | 223.29 | 17.58 | 1118 | 229.10 | 18.95 |
| Heifer 2 | 251 | 233.31 | 20.15 | 299 | 226.91 | 19.22 | 374 | 226.72 | 18.82 | 924 | 228.57 | 19.36 |
| Heifer 3 | 67 | 224.42 | 18.84 | 342 | 233.79 | 19.37 | 194 | 229.71 | 19.22 | 603 | 231.44 | 19.29 |
| Overall | 541 | 233.28 | 19.47 | 1109 | 230.83 | 19.57 | 995 | 225.84 | 18.40 | 2645 | 229.45 | 19.17 |
| lnCH4 | ||||||||||||
| Heifer 1 | 243 | 5.60 | 5.13 | 484 | 5.58 | 4.90 | 424 | 5.54 | 4.87 | 1151 | 5.57 | 4.96 |
| Heifer 2 | 253 | 5.59 | 4.50 | 312 | 5.57 | 5.34 | 357 | 5.54 | 4.78 | 922 | 5.56 | 4.91 |
| Heifer 3 | 62 | 5.54 | 4.60 | 345 | 5.56 | 4.53 | 206 | 5.59 | 5.08 | 613 | 5.57 | 4.74 |
| Overall | 558 | 5.59 | 4.80 | 1141 | 5.57 | 4.91 | 987 | 5.55 | 4.89 | 2686 | 5.57 | 4.89 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niero, G.; Cendron, F.; Penasa, M.; De Marchi, M.; Cozzi, G.; Cassandro, M. Repeatability and Reproducibility of Measures of Bovine Methane Emissions Recorded using a Laser Detector. Animals 2020, 10, 606. https://doi.org/10.3390/ani10040606
Niero G, Cendron F, Penasa M, De Marchi M, Cozzi G, Cassandro M. Repeatability and Reproducibility of Measures of Bovine Methane Emissions Recorded using a Laser Detector. Animals. 2020; 10(4):606. https://doi.org/10.3390/ani10040606
Chicago/Turabian StyleNiero, Giovanni, Filippo Cendron, Mauro Penasa, Massimo De Marchi, Giulio Cozzi, and Martino Cassandro. 2020. "Repeatability and Reproducibility of Measures of Bovine Methane Emissions Recorded using a Laser Detector" Animals 10, no. 4: 606. https://doi.org/10.3390/ani10040606





