Effects on the Ileal Microbiota of Phosphorus and Calcium Utilization, Bird Performance, and Gender in Japanese Quail
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Sample Collection, DNA Extraction, and Illumina Library Preparation
2.3. Samples Grouping
2.4. Bioinformatics and Stratistical Analysis
3. Results and Discussion
3.1. Effect of PU, CaU, and Animal Performance on Microbial Distribution
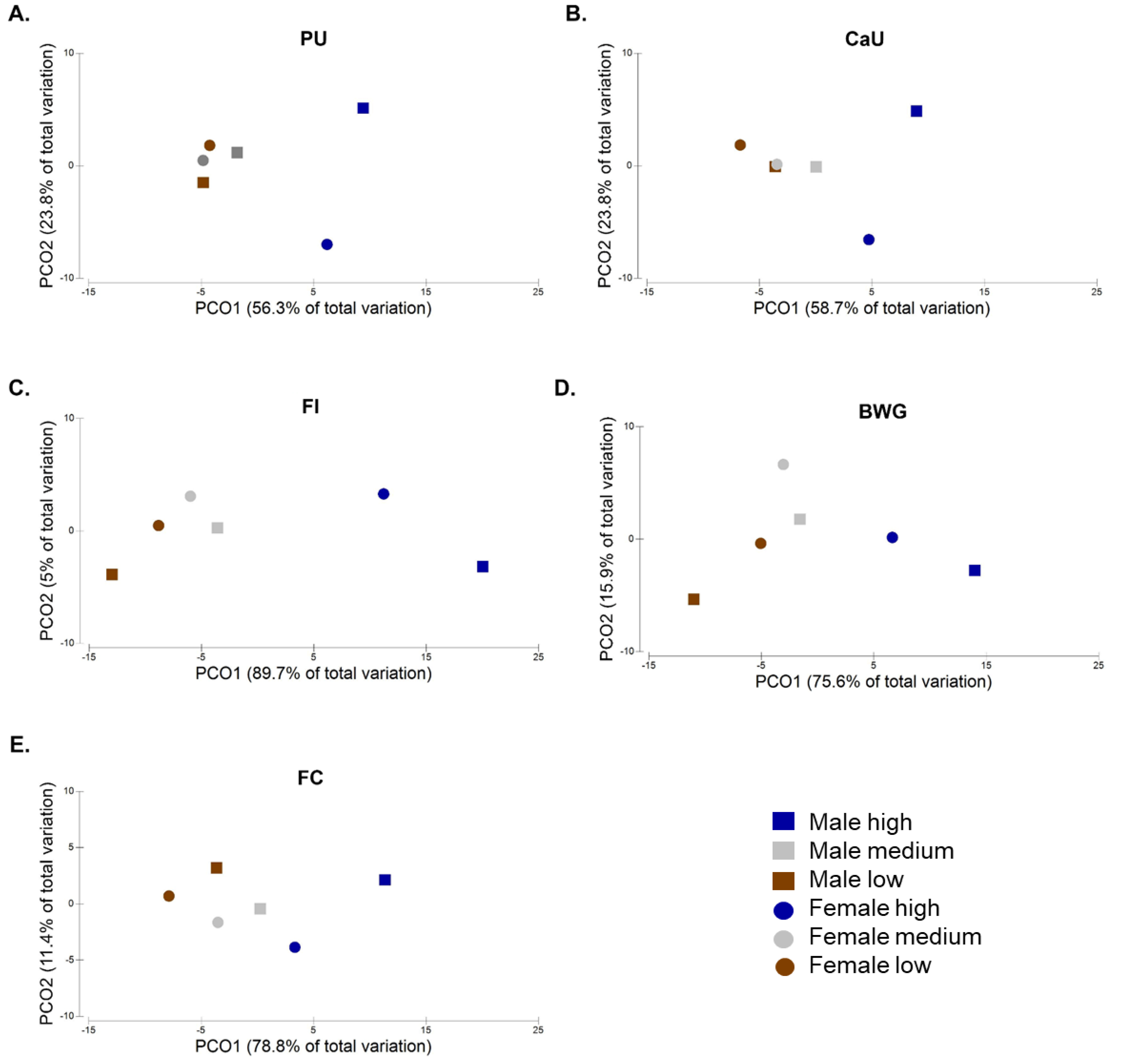
3.2. Gender Effects on Microbiota, PU, CaU, FI, BWG, and FC
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mills, A. The behavior of the Japanese or domestic quail Coturnix japonica. Neurosci. Biobehav. Rev. 1997, 21, 261–281. [Google Scholar] [CrossRef]
- Padgett, C.A.; Ivey, W.D. Coturnix quail as a laboratory research animal. Science 1959, 129, 267–268. [Google Scholar] [CrossRef] [PubMed]
- Minvielle, F.; Gourichon, D.; Ito, S.; Inoue-Murayama, M.; Rivière, S. Effects of the dominant lethal yellow mutation on reproduction, growth, feed consumption, body temperature, and body composition of the Japanese quail. Poult. Sci. 2007, 86, 1646–1650. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.M.; Bennett, D.C.; Mills, A.D. The Japanese Quail. In The UFAW Handbook on the Care and Management of Laboratory and Other Research Animals, 8th ed.; Hubrecht, R., Kirkwood, J.K., Eds.; Wiley-Blackwell: Chichester/West Sussex, UK; Ames, IA, USA, 2010; pp. 655–673. [Google Scholar]
- Rodehutscord, M.; Dieckmann, A. Comparative studies with three-week-old chickens, turkeys, ducks, and quails on the response in phosphorus utilization to a supplementation of monobasic calcium phosphate. Poult. Sci. 2005, 84, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, N.; Hughes, R.J.; Aspden, W.J.; Chapman, J.; Moore, R.J.; Stanley, D. The gastrointestinal tract microbiota of the Japanese quail, Coturnix japonica. Appl. Microbiol. Biotechnol. 2016, 100, 4201–4209. [Google Scholar] [CrossRef] [PubMed]
- Kohl, K.D.; Amaya, J.; Passement, C.A.; Dearing, M.D.; McCue, M.D. Unique and shared responses of the gut microbiota to prolonged fasting: A comparative study across five classes of vertebrate hosts. FEMS Microbiol. Ecol. 2014, 90, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Bennett, D.C.; Tun, H.M.; Kim, J.-E.; Cheng, K.M.; Zhang, H.; Leung, F.C. The effect of diet and host genotype on ceca microbiota of Japanese quail fed a cholesterol enriched diet. Front. Microbiol. 2015, 6, 1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Wang, G.; Siegel, P.; He, C.; Wang, H.; Zhao, W.; Zhai, Z.; Tian, F.; Zhao, J.; Zhang, H.; et al. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci. Rep. 2013, 3, 1163. [Google Scholar] [CrossRef]
- Grond, K.; Sandercock, B.K.; Jumpponen, A.; Zeglin, L.H. The avian gut microbiota: Community, physiology and function in wild birds. J. Avian Biol. 2018, 49, e01788. [Google Scholar] [CrossRef] [Green Version]
- Ptak, A.; Bedford, M.R.; Swiatkiewicz, S.; Zyla, K.; Jozefiak, D. Phytase modulates ileal microbiota and enhances growth performance of the broiler chickens. PLoS ONE 2015, 10, e0119770. [Google Scholar] [CrossRef] [Green Version]
- Borda-Molina, D.; Vital, M.; Sommerfeld, V.; Rodehutscord, M.; Camarinha-Silva, A. Insights into Broilers’ Gut Microbiota Fed with Phosphorus, Calcium, and Phytase Supplemented Diets. Front. Microbiol. 2016, 7, 2033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, P.; Piepho, H.-P.; Rodehutscord, M.; Bennewitz, J. Inferring relationships between Phosphorus utilization, feed per gain, and bodyweight gain in an F2 cross of Japanese quail using recursive models. Poult. Sci. 2016, 95, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Gesellschaft für Ernährungsphysiologie (GfE). Proceedings Society Nutrition Physiologie: Empfelungen zur Energie- und Nährstoffversorgung von Mastputen; DLG Verlag: Frankfurt am Main, Germany, 2004. [Google Scholar]
- Künzel, S.; Bennewitz, J.; Rodehutscord, M. Genetic parameters for bone ash and phosphorus utilization in an F2 cross of Japanese quail. Poult. Sci. 2019, 98, 4369–4372. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Arriaga, A.; Baumann, A.; Witte, O.W.; Frahm, C.; Bergheim, I.; Camarinha-Silva, A. Changes in Oral Microbial Ecology of C57BL/6 Mice at Different Ages Associated with Sampling Methodology. Microorganisms 2019, 7, 283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etchebehere, C.; Tiedje, J. Presence of two different active nirS nitrite reductase genes in a denitrifying Thauera sp. from a high-nitrate-removal-rate reactor. Appl. Environ. Microbiol. 2005, 71, 5642–5645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, D.J. Nucleic acid Techniques in Bacterial Systematics: 16S/23S rRNA Sequencing; Wiley & Sons: Chichester, UK, 1991. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Argüello, H.; Estellé, J.; Zaldívar-López, S.; Jiménez-Marín, Á.; Carvajal, A.; López-Bascón, M.A.; Crispie, F.; O’Sullivan, O.; Cotter, P.D.; Priego-Capote, F.; et al. Early Salmonella Typhimurium infection in pigs disrupts Microbiome composition and functionality principally at the ileum mucosa. Sci. Rep. 2018, 8, 7788. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Clarke, K.R.; Warwick, R.M. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation; PRIMER-E: Plymouth, UK, 2001. [Google Scholar]
- Reda, F.M.; Alagawany, M.; Mahmoud, H.K.; Mahgoub, S.A.; Elnesr, S.S. Use of red pepper oil in quail diets and its effect on performance, carcass measurements, intestinal microbiota, antioxidant indices, immunity and blood constituents. Animal 2020, 14, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tun, H.M.; Leung, F.C.; Bennett, D.C.; Zhang, H.; Cheng, K.M. Interaction of genotype and diet on small intestine microbiota of Japanese quail fed a cholesterol enriched diet. Sci. Rep. 2018, 8, 2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borda-Molina, D.; Seifert, J.; Camarinha-Silva, A. Current perspectives of the chicken gastrointestinal tract and its microbiome. Comput. Struct. Biotechnol. J. 2018, 16, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chaudhry, M.T.; Zhao, D.; Lin, T.; Tian, Y.; Fu, J. Heat shock protein 70 protects the quail cecum against oxidant stress, inflammatory injury, and microbiota imbalance induced by cold stress. Poult. Sci. 2019, 98, 5432–5445. [Google Scholar] [CrossRef]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Kalsum, U.; Soetanto, H.A.; Sjofjan, O. Influence of a Probiotic Containing Lactobacillus fermentum on the Laying Performance and Egg Quality of Japanese Quails. Int. J. Poult. Sci. 2012, 11, 311–315. [Google Scholar] [CrossRef] [Green Version]
- Samli, H.E.; Senkoylu, N.; Koc, F.; Kanter, M.; Agma, A. Effects of Enterococcus faecium and dried whey on broiler performance, gut histomorphology and intestinal microbiota. Arch. Anim. Nutr. 2007, 61, 42–49. [Google Scholar] [CrossRef]
- Lauková, A.; Guba, P.; Nemcová, R.; Vasilková, Z. Reduction of Salmonella in gnotobiotic Japanese quails caused by the enterocin A-producing EK13 strain of Enterococcus Faecium. Vet. Res. Commun. 2003, 27, 275–280. [Google Scholar] [CrossRef]
- Richards-Rios, P.; Fothergill, J.; Bernardeau, M.; Wigley, P. Development of the Ileal Microbiota in Three Broiler Breeds. Front. Vet. Sci. 2020, 7, e91941. [Google Scholar] [CrossRef] [Green Version]
- Danzeisen, J.L.; Calvert, A.J.; Noll, S.L.; McComb, B.; Sherwood, J.S.; Logue, C.M.; Johnson, T.J. Succession of the turkey gastrointestinal bacterial microbiome related to weight gain. PeerJ 2013, 1, e237. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Youmans, B.P.; Noll, S.; Cardona, C.; Evans, N.P.; Karnezos, T.P.; Ngunjiri, J.M.; Abundo, M.C.; Lee, C.-W. A Consistent and Predictable Commercial Broiler Chicken Bacterial Microbiota in Antibiotic-Free Production Displays Strong Correlations with Performance. Appl. Environ. Microbiol. 2018, 84, e00362-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.-L.; Wang, J.; Zhang, H.-J.; Wu, S.-G.; Hui, Q.-R.; Yang, C.-B.; Fang, R.-J.; Qi, G.-H. Intestinal Morphologic and Microbiota Responses to Dietary Bacillus spp. in a Broiler Chicken Model. Front. Physiol. 2018, 9, 1968. [Google Scholar] [CrossRef] [PubMed]
- Hmani, H.; Daoud, L.; Jlidi, M.; Jalleli, K.; Ben Ali, M.; Hadj Brahim, A.; Bargui, M.; Dammak, A.; Ben Ali, M. A Bacillus subtilis strain as probiotic in poultry: Selection based on in vitro functional properties and enzymatic potentialities. J. Ind. Microbiol. Biotechnol. 2017, 44, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
- Prakash, J.; Arora, N.K. Phosphate-solubilizing Bacillus sp. enhances growth, phosphorus uptake and oil yield of Mentha arvensis L. 3 Biotech 2019, 9, 126. [Google Scholar]
- Saeid, A.; Prochownik, E.; Dobrowolska-Iwanek, J. Phosphorus Solubilization by Bacillus Species. Molecules 2018, 23, 2897. [Google Scholar] [CrossRef] [Green Version]
- Rezaei, M.; Karimi Torshizi, M.A.; Wall, H.; Ivarsson, E. Body growth, intestinal morphology and microflora of quail on diets supplemented with micronised wheat fibre. Br. Poult. Sci. 2018, 59, 422–429. [Google Scholar] [CrossRef]
- Beck, P.; Rodehutscord, M.; Bennewitz, J.; Bessei, W. A pilot study of the genetic variation of phosphorus utilization in young Japanese quail (Coturnix japonica). Poult. Sci. 2014, 93, 1916–1921. [Google Scholar] [CrossRef] [Green Version]
- Parois, S.; Calandreau, L.; Kraimi, N.; Gabriel, I.; Leterrier, C. The influence of a probiotic supplementation on memory in quail suggests a role of gut microbiota on cognitive abilities in birds. Behav. Brain Res. 2017, 331, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Kraimi, N.; Calandreau, L.; Biesse, M.; Rabot, S.; Guitton, E.; Velge, P.; Leterrier, C. Absence of Gut Microbiota Reduces Emotional Reactivity in Japanese Quails (Coturnix japonica). Front. Physiol. 2018, 9, 603. [Google Scholar] [CrossRef]
- Capcarova, M.; Kalafova, A.; Lajdova, Z.; Schwarzova, M.; Zbynovska, K.; Hrncar, C.; Hanusova, E.; Brunaiova, Z.; Bielik, P. Effectiveness of non-antibiotic stimulators in Japanese quail diet: Gender comparison and economical annex. Biologia 2017, 72, 255. [Google Scholar] [CrossRef]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken Gut Microbiota: Importance and Detection Technology. Front. Vet. Sci. 2018, 5, 254. [Google Scholar] [CrossRef] [PubMed]
- Crisol-Martínez, E.; Moreno-Moyano, L.T.; Wilkinson, N.; Prasai, T.; Brown, P.H.; Moore, R.J.; Stanley, D. A low dose of an organophosphate insecticide causes dysbiosis and sex-dependent responses in the intestinal microbiota of the Japanese quail (Coturnix japonica). PeerJ 2016, 4, e2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apajalahti, J.; Kettunen, A.; Graham, H. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. World’s Poult. Sci. J. 2004, 60, 223–232. [Google Scholar] [CrossRef]
- Wilkinson, N.; Hughes, R.J.; Bajagai, Y.S.; Aspden, W.J.; Hao Van, T.T.; Moore, R.J.; Stanley, D. Reduced environmental bacterial load during early development and gut colonisation has detrimental health consequences in Japanese quail. Heliyon 2020, 6, e03213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witzig, M.; Carminha-Silva, A.; Camarinha da Silva, A.; Green-Engert, R.; Hoelzle, K.; Zeller, E.; Seifert, J.; Hoelzle, L.E.; Rodehutscord, M. Spatial Variation of the Gut Microbiota in Broiler Chickens as Affected by Dietary Available Phosphorus and Assessed by T-RFLP Analysis and 454 Pyrosequencing. PLoS ONE 2015, 10, e0143442. [Google Scholar]
- Farrow, J.A.E.; Kruze, J.; Phillips, B.A.; Bramley, A.J.; Collins, M.D. Taxonomic studies on Streptococcus bovis and Streptococcus equinus: Description of Streptococcus alactolyticus sp. nov. and Streptococcus saccharolyticus sp. nov. Syst. Appl. Microbiol. 1984, 5, 467–482. [Google Scholar] [CrossRef]
- Yang, W.-Y.; Lee, Y.; Lu, H.; Chou, C.-H.; Wang, C. Analysis of gut microbiota and the effect of lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model. PLoS ONE 2019, 14, e0205784. [Google Scholar] [CrossRef] [Green Version]
- Dipineto, L.; Russo, T.P.; Gargiulo, A.; Borrelli, L.; de Luca Bossa, L.M.; Santaniello, A.; Buonocore, P.; Menna, L.F.; Fioretti, A. Prevalence of enteropathogenic bacteria in common quail (Coturnix coturnix). Avian Pathol. 2014, 43, 498–500. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borda-Molina, D.; Roth, C.; Hérnandez-Arriaga, A.; Rissi, D.; Vollmar, S.; Rodehutscord, M.; Bennewitz, J.; Camarinha-Silva, A. Effects on the Ileal Microbiota of Phosphorus and Calcium Utilization, Bird Performance, and Gender in Japanese Quail. Animals 2020, 10, 885. https://doi.org/10.3390/ani10050885
Borda-Molina D, Roth C, Hérnandez-Arriaga A, Rissi D, Vollmar S, Rodehutscord M, Bennewitz J, Camarinha-Silva A. Effects on the Ileal Microbiota of Phosphorus and Calcium Utilization, Bird Performance, and Gender in Japanese Quail. Animals. 2020; 10(5):885. https://doi.org/10.3390/ani10050885
Chicago/Turabian StyleBorda-Molina, Daniel, Christoph Roth, Angélica Hérnandez-Arriaga, Daniel Rissi, Solveig Vollmar, Markus Rodehutscord, Jörn Bennewitz, and Amélia Camarinha-Silva. 2020. "Effects on the Ileal Microbiota of Phosphorus and Calcium Utilization, Bird Performance, and Gender in Japanese Quail" Animals 10, no. 5: 885. https://doi.org/10.3390/ani10050885




