The Genetic Diversity and Population Genetic Structure of the Red Panda, Ailurus fulgens, in Zoos in China
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Sampling
2.2. DNA Extraction
2.3. Polymerase Chain Reaction and Genotyping
2.4. Statistical Method
3. Results
3.1. Genetic Diversity of Populations
3.2. Hardy–Weinberg Equilibrium Test
3.3. Genetic Differentiation between Pairs of Populations
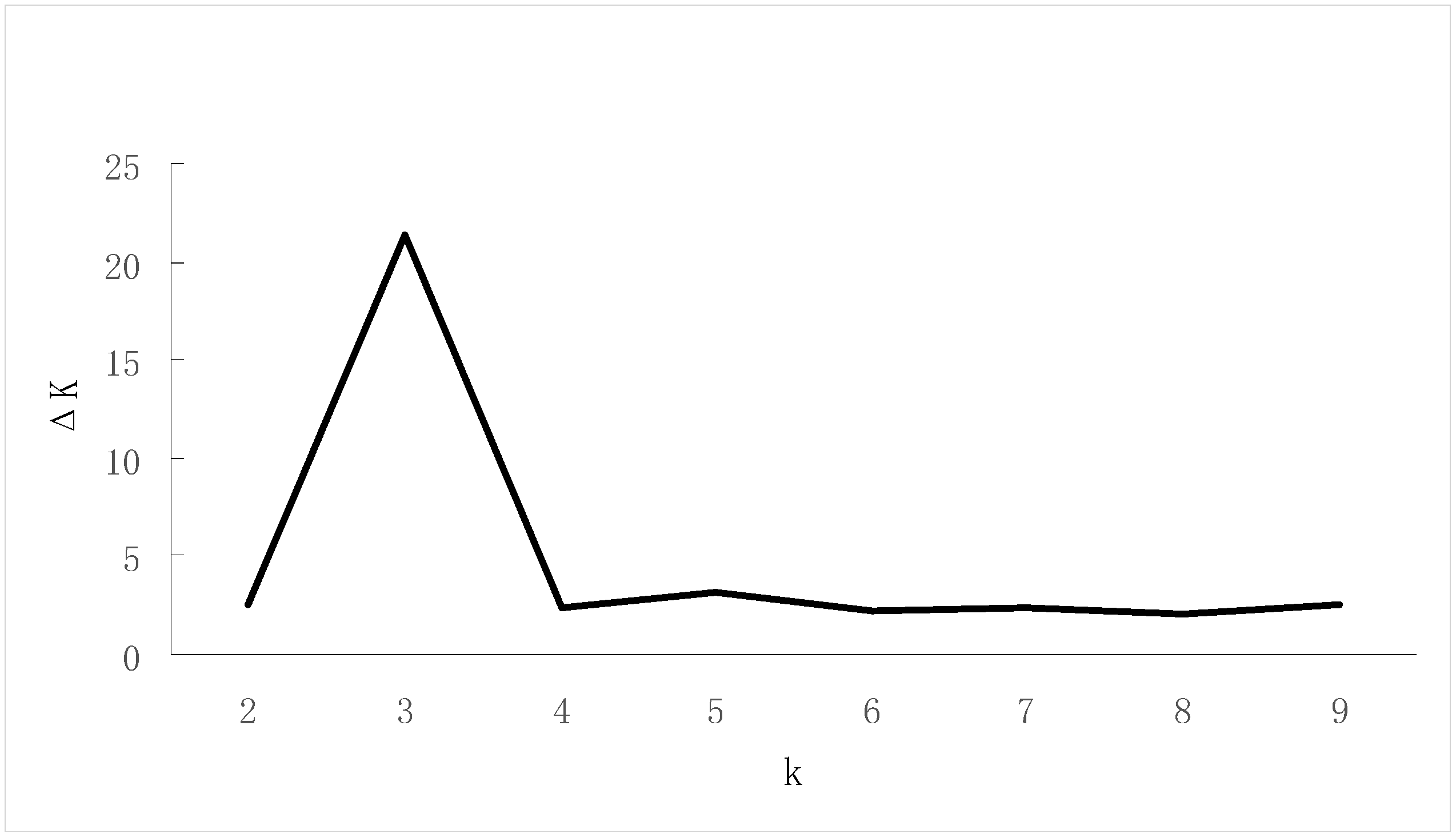
3.4. Genetic Structure Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Su, B.; Fu, Y.; Wang, Y.X.; Chakraborty, R. Genetic diversity and population history of the red panda (Ailurus fulgens) as inferred from mitochondrial DNA sequence variations. Mol. Biol. Evol. 2001, 18, 1070–1076. [Google Scholar] [CrossRef] [Green Version]
- Wei, F.; Feng, Z.; Wang, Z.; Hu, J. Current distribution, status and conservation of wild red pandas Ailurus fulgens in China. Biol. Conserv. 1997, 89, 285–291. [Google Scholar] [CrossRef]
- Kumar, A.; Rai, U.; Roka, B.; Jha, A.K.; Reddy, P.A. Genetic assessment of captive red panda (Ailurus fulgens) population. SpringerPlus. 2016, 5, 1750. [Google Scholar] [CrossRef] [Green Version]
- Thapa, A.; Hu, Y.; Wei, F. The endangered red panda (Ailurus fulgens): Ecology and conservation approaches across the entire range. Biol. Conserv. 2018, 220, 112–121. [Google Scholar] [CrossRef]
- Yue, T.X. Studies and questions of biological diversity. Acta Ecol. Sin. 2001, 21, 462–467. [Google Scholar]
- Hu, H.G.; Shen, X.Y. The history and status of captive red panda breeding in China. Wild Anim. 1997, 18, 16–18. [Google Scholar]
- Hu, Y.; Guo, Y.U.; Qi, D.; Zhan, X.; Wu, H.U.A.; Bruford, M.W.; Wei, F. Genetic structuring and recent demographic history of red pandas (Ailurus fulgens) inferred from microsatellite and mitochondrial DNA. Mol. Ecol. 2011, 20, 2662–2675. [Google Scholar] [CrossRef]
- Liang, X. Construct the microsatellite libraries from red panda (Ailurus fulgens) and identified the loci used for analysis. Master’s Thesis, Sichuan University, Chengdu, China, 2007. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Liu, Z.J.; Zhang, B.W.; Wei, F.W.; Li, M. Isolation and characterization of microsatellite loci for the red panda (Ailurus fulgens). Mol. Ecol. Notes 2005, 5, 27–29. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, X.; Zhang, Z.; Shen, F.; Zhang, W.; Wei, K.; Yang, Z.; Hou, R.; Yue, B.; Kamata, H.; et al. Identification and characterization of ten polymorphic microsatellite loci in red panda Ailurus fulgens. Conserv. Genet. 2008, 9, 787–790. [Google Scholar] [CrossRef]
- Wu, H.; Zhan, X.J.; Guo, Y.; Zhang, Z.; Zhu, L.; Yan, L.; Li, M.; Wei, F.-W. Isolation and characterization of 12 novel microsatellite loci for the red panda (Ailurus fulgens). Conserv. Genet. 2009, 10, 523–525. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47. [Google Scholar] [CrossRef] [Green Version]
- Goudet, J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Updated from Goudet (1995). Available online: https://www.scienceopen.com/document?vid=b577cf2d-d522-4831-a4a7-d05d435e72cf (accessed on 8 June 2020).
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- You, Y.Y. Influence on endemic bat Myotis davidii population size and genetic richenss by the habit cave interference. J. Biol. 2013, 32, 314–331. [Google Scholar]
- Yan, L.; Huang, Y.; Zhang, B.; Zhang, S.; Zhang, H.; Wei, F.; Wang, P. Status and prognosis of genetic diversity in captive giant pandas (Ailuropoda melanoleuca) in Wolong. Acta Thériol. Sin. 2006, 26, 317–324. [Google Scholar]
- Xiu, Y.F.; Chen, Y.C.; Shao, L.P.; Xu, S.H. Rearing and Reproduction of Lesser Panda in Fuzhou Area. Sichuan J. Zool. 2007, 25, 875–878. [Google Scholar]
- Nechtelberger, D.; Kaltwasser, C.; Stur, I.; Meyer, J.N.; Brem, G.; Mueller, M.; Mueller, S. DNA microsatellite analysis for parentage control in austrian pigs. Anim. Biotechnol. 2001, 12, 141–144. [Google Scholar] [CrossRef]
- Ardren, W.R.; Borer, S.; Thrower, F.; Joyce, J.E.; Kapuscinski, A.R. Inheritance of 12 microsatellite loci in Oncorhynchus mykiss. J. Hered. 1999, 90, 529–536. [Google Scholar] [CrossRef] [Green Version]
- Xiu, Y.F.; Li, B.C.; Chen, Y.S.; Zhang, Y.N.; Xu, S.H. Paternity identification of red panda using microsatellite genotyping. Acta Thériol. Sin. 2015, 35, 55–64. [Google Scholar]
- Wright, S. Varisbility Within and Among Natural Populations. In Evolution and the Genetics of Populations; University of Chicago Press: Chicago, IL, USA, 1978; Volume 4. [Google Scholar]
- Wang, F.; Peng, Z.-X.; Zhang, J.-G.; Tian, L.; Han, H.-B.; Cao, Z.-C.; Zhang, Z.-H.; Shen, F.-J.; Li, D.-S.; Zhang, H.-M.; et al. Studies on genetic diversity of captive giant pandas (Ailuropoda melanoleuca) based on microsatellite DNA markers. Prog. Biochem. Biophys. 2007, 34, 1279–1287. [Google Scholar]

| Loci | Reference | GenBank Accession No. | Primers Sequences (5′–3′) | Repeat Motif | Labelling Dye | Size Range (bp) | Ta (°C) |
|---|---|---|---|---|---|---|---|
| RP-1 | [10] | AY685403 | F:CGCCCAGGTACCCTAGAGT | (GT)5AT(GT)10 | FITC | 160–198 | 58 |
| R:TCCCCACGTTCACTGCAGCATTATC | |||||||
| RP-11 | [10] | AY685406 | F:TGAATGTTGCCTTGCTCT | (CA)12 | FAM | 101–127 | 51 |
| R:CACCACCTCTTACTGTTCTC | |||||||
| RP-101 | [10] | AY685406 | F:ACAGACTGAAAAGGCTTAACAGAGT | (AG)20 | HEX | 155–173 | 56 |
| R:CGGTCCATGGTATTCTCCTGTAG | |||||||
| RP-102 | [10] | AY685407 | F:ATGCAGAAGAGAATGGAAGCCTGG | (CG)4(TG)10 | HEX | 199–209 | 57 |
| R:TTCCTGGCAACGATTTCATCCTCAA | |||||||
| RP-108 | [10] | AY60885 | F:GTCTCCTCTAACAGCCCACAC | (TG)3(CA)3CTTA(CA)16 | TAMRA | 217–265 | 58 |
| R:GAGGCCACTCTCAACTTTAGTAGAA | |||||||
| RP-133 | [10] | AY685409 | F:GCAGGAAGAAGAGGTACTGGTTTCA | (AC)16 | TAMRA | 205–233 | 56 |
| R:AACAAGATGCCAGGAAGATACTTTC | |||||||
| RP-137 | [10] | AY685410 | F:CACCGTACCCGAGACACCAA | (AC)16(AG)12 | FAM | 141–153 | 52 |
| R:AAAGAAGAGAAAGTGAAGTGGCAGC | |||||||
| Aifu-1 | [8] | EF408640 | F:CCTGCATCAGACTCAGCA | (ATAG)12 | FAM | 140–176 | 58 |
| R:GGTATCAGACGTGGGAACTA | |||||||
| Aifu-2 | [8] | EF408641 | F:GACCCAGCCCTAACTCAAA | (CTAC)11 | HEX | 125–141 | 57 |
| R:CCTGCATTAGGCTCCACA | |||||||
| Aifu-5 | [8] | EF408644 | F:GAATAATGAGCTTGCCTTCC | (GATA)13 | TAMRA | 336–352 | 55 |
| R:TTGACATTGGCTATGTGAACA | |||||||
| Aifu-7 | [8] | EF408646 | F:CCTGCATCAGACTCAGCA | (GATA)11 | HEX | 221–249 | 60 |
| R:CTGGCTTGCAGACAGGAAAT | |||||||
| Aifu-8 | [8] | EF408647 | F:TTTACAAAGCAGAGCGTC | (GATA)15 | FAM | 123–171 | 59 |
| R:AAATCCTGTCGAACCATG | |||||||
| Aifu-25 | [8] | EF408651 | F:AATTGCATGAGCCAGTTC | (CTAT)15 | HEX | 153–205 | 54 |
| R:GCCAGGGTTTTACCAGAG | |||||||
| Aifu-27 | [8] | EF408655 | F:CTCAGAATCTTTCATTGCCA | (CTAT)13CCACA(GT)15 | TAMRA | 255–289 | 54 |
| R:TTCCTCAATCCTCTGTTCAA | |||||||
| Aifu-12 | [11] | EF620036 | F:TTTGCCAGTAAGCACCCG | (CA)19 | TAMRA | 259–269 | 58 |
| R:TGGAACTCAGAGGAGCAGTC | |||||||
| Aifu-15 | [11] | EF620039 | F:AGTACCAGCAGTAGGATAGCA | (CA)3CG(CA)6 | FAM | 120–150 | 58 |
| R:TAAGAGCACTCAGAGGGAAAC | |||||||
| Rp6 | [12] | EU487250 | F:ACTGAGGCGAAGAAAGGAGC | (CA)13 | HEX | 154–166 | 56 |
| R:CATGGGCATTGAAGATGGTG | |||||||
| Rp9 | [12] | EU487208 | F:ACAAACTGGAATGTAAAGG | (CA)20 | TAMRA | 125–143 | 58 |
| R:CACATGCTTATGTTATAGGG | |||||||
| RP13 | [12] | EU487212 | F:TCCCTTACGCTTCCTCCTTT | (GT)18 | HEX | 126–224 | 52 |
| R:GCAGGCGGAGAATTGGTTGG |
| Loci | Number of Alleles (NA) | Expected Heterozygosity (HE) | Observed Heterozygosity (Ho) | Polymorphism Information Content (PIC) | Fixation Indices of the Total Population (FIT) | Genetic Differentiation Index (FST) | Inbreeding Coefficient within the Subpopulation (FIS) |
|---|---|---|---|---|---|---|---|
| RP-1 | 11 | 0.568 | 0.547 | 0.553 | 0.019 | 0.062 ** | −0.046 |
| RP-101 | 8 | 0.853 | 0.866 | 0.829 | −0.018 | 0.037 ** | −0.057 |
| RP-102 | 5 | 0.659 | 0.600 | 0.606 | 0.099 * | 0.09 ** | 0.011 |
| RP-108 | 13 | 0.852 | 0.703 | 0.837 | 0.138 ** | 0.032 ** | 0.110 ** |
| RP-133 | 12 | 0.814 | 0.844 | 0.787 | −0.028 | 0.055 ** | −0.086 |
| RP-137 | 7 | 0.713 | 0.595 | 0.676 | 0.155 ** | 0.105 ** | 0.058 |
| Aifu-25 | 10 | 0.769 | 0.786 | 0.739 | −0.024 | 0.037 ** | −0.064 |
| Aifu-27 | 18 | 0.901 | 0.926 | 0.889 | −0.024 | 0.031 ** | −0.056 |
| Aifu-39 | 6 | 0.528 | 0.513 | 0.469 | 0.015 | 0.038 ** | −0.026 |
| Aifu-1 | 9 | 0.827 | 0.867 | 0.848 | 0.049 * | 0.035 ** | 0.015 |
| Aifu-2 | 5 | 0.571 | 0.525 | 0.515 | 0.120 * | 0.115 ** | 0.005 |
| Aifu-5 | 5 | 0.732 | 0.762 | 0.681 | −0.046 | 0.033 * | −0.082 |
| Aifu-7 | 11 | 0.776 | 0.519 | 0.762 | 0.269 ** | 0.067 ** | 0.217 ** |
| Aifu-8 | 12 | 0.854 | 0.835 | 0.845 | 0.038 | 0.056 ** | −0.018 |
| Aifu-12 | 6 | 0.730 | 0.875 | 0.681 | −0.209 | 0.04 ** | −0.259 |
| Aifu-15 | 7 | 0.661 | 0.602 | 0.599 | 0.111 | 0.034 * | 0.08 |
| Rp6 | 6 | 0.601 | 0.625 | 0.531 | −0.075 | 0.03 ** | −0.108 |
| Rp9 | 10 | 0.779 | 0.867 | 0.751 | 0.115 ** | 0.091 ** | 0.027 |
| RP11 | 14 | 0.878 | 0.809 | 0.856 | 0.097 * | 0.067 ** | 0.032 |
| All loci | 175 | 0.049 ** | 0.055 ** | −0.006 | |||
| Mean ± SD | 9.210 ± 3.6 | 0.730 ± 0.115 | 0.719 ± 0.145 | 0.708 ± 0.131 |
| Locus | * Chengdu | * Pixian | * Kunming | Fuzhou | Wuxi | Shanghai | Beijing | Panyu | Guangzhou | Chongqi | Dazu | Mianning | Huangshan | Wenling |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aifu-1 | 0.852 | 0.811 | 0.786 | 0.878 | 0.865 | 0.875 | 0.727 | 0.833 | 0.769 | 0.773 | 0.833 | 0.765 | 0.857 | 0.833 |
| Aifu-2 | 0.720 | 0.742 | 0.464 | 0.408 | 0.697 | 0.739 | 0.439 | 0.625 | 0.560 | 0.409 | 0.697 | 0.451 | 0.495 | 0.621 |
| Aifu-5 | 0.787 | 0.721 | 0.250 | 0.688 | 0.783 | 0.654 | 0.714 | 0.767 | 0.703 | 0.689 | 0.750 | 0.717 | 0.833 | 0.712 |
| Aifu-7 | 0.606 | 0.774 | 0.679 | 0.805 | 0.662 | 0.882 | 0.511 | 0.517 | 0.747 | 0.682 | 0.429 | 0.846 | 0.788 | 0.733 |
| Aifu-8 | 0.833 | 0.774 | 0.893 | 0.864 | 0.805 | 0.556 | 0.818 | 0.858 | 0.857 | 0.712 | 0.848 | 0.941 | 0.846 | 0.879 |
| Aifu-25 | 0.803 | 0.884 | 0.785 | 0.784 | 0.706 | 0.732 | 0.712 | 0.700 | 0.697 | 0.667 | 0.733 | 0.692 | 0.758 | 0.833 |
| Aifu-27 | 0.874 | 0.948 | 0.893 | 0.885 | 0.784 | 0.955 | 0.864 | 0.875 | 0.933 | 0.879 | 0.848 | 0.950 | 0.955 | 0.844 |
| Aifu-12 | 0.862 | 0.816 | 0.750 | 0.763 | 0.827 | 0.582 | 0.682 | 0.658 | 0.626 | 0.758 | 0.644 | 0.642 | 0.511 | 0.712 |
| Aifu-15 | 0.667 | 0.784 | 0.893 | 0.585 | 0.662 | 0.442 | 0.803 | 0.700 | 0.788 | 0.644 | 0.758 | 0.660 | 0.538 | 0.803 |
| Aifu-39 | 0.636 | 0.537 | 0.857 | 0.508 | 0.500 | 0.621 | 0.409 | 0.517 | 0.143 | 0.318 | 0.591 | 0.621 | 0.582 | 0.652 |
| Mean | 0.764 | 0.779 | 0.725 | 0.717 | 0.729 | 0.704 | 0.668 | 0.705 | 0.682 | 0.653 | 0.713 | 0.728 | 0.716 | 0.762 |
| Populations | No. of Individuals | MNA | HO | HE |
|---|---|---|---|---|
| Qionglai | 26 | 6.2 | 0.743 | 0.688 |
| Liangshan | 28 | 5.8 | 0.700 | 0.704 |
| Xiaoxiangling | 27 | 5.8 | 0.564 | 0.634 |
| Gaoligong | 20 | 7.1 | 0.712 | 0.732 |
| Tibet | 4 | 4.6 | 0.722 | 0.770 |
| The wild population | 105 | 9.2 | 0.679 | 0.719 |
| Fuzhou | 41 | 7.1 | 0.732 | 0.722 |
| Wuxi | 11 | 5.2 | 0.711 | 0.647 |
| Shanghai | 9 | 5.0 | 0.693 | 0.747 |
| Mianning | 9 | 4.0 | 0.715 | 0.734 |
| Panyu | 8 | 5.0 | 0.693 | 0.746 |
| Guangzhou | 7 | 4.5 | 0.669 | 0.684 |
| Huangshan | 7 | 3.8 | 0.702 | 0.631 |
| Beijing | 6 | 4.1 | 0.683 | 0.744 |
| Chongqi | 6 | 4.2 | 0.673 | 0.644 |
| Dazu | 6 | 3.9 | 0.749 | 0.734 |
| Wenling | 6 | 3.9 | 0.740 | 0.782 |
| The captive population | 116 | 9.3 | 0.719 | 0.730 |
| Population | Number of Individual | Mean Number of Alleles (MNA) | Mean Allelic Richness (MAR) | Expected Heterozygosity (HE) | Observed Heterozygosity (Ho) | Inbreeding Coefficient within the Subpopulation (FIS) | Hardy-Weinberg Equilibrium (p) |
|---|---|---|---|---|---|---|---|
| Fuzhou | 41 | 7.11 | 3.913 | 0.722 | 0.732 | −0.033 | 0.0005 * |
| Wuxi | 11 | 5.21 | 3.751 | 0.647 | 0.711 | 0.088 * | 0.0019 * |
| Shanghai | 9 | 5.00 | 3.767 | 0.747 | 0.693 | −0.083 | 0.5411 |
| Beijing | 6 | 4.05 | 3.505 | 0.744 | 0.683 | −0.173 * | 0.7836 |
| Panyu | 8 | 4.95 | 3.837 | 0.746 | 0.693 | 0.093 | 0.1594 |
| Guangzhou | 7 | 4.53 | 3.748 | 0.684 | 0.669 | −0.013 | 0.7825 |
| Chongqi | 6 | 4.16 | 3.573 | 0.644 | 0.673 | 0.062 | 0.5984 |
| Dazu | 6 | 4.37 | 3.908 | 0.734 | 0.749 | 0.015 | 0.4263 |
| Mianning | 9 | 5.42 | 4.026 | 0.728 | 0.715 | 0.001 | 0.5719 |
| Huangshan | 7 | 4.58 | 3.797 | 0.631 | 0.702 | 0.111 * | 0.5751 |
| Wenling | 6 | 4.53 | 3.903 | 0.782 | 0.740 | −0.067 | 0.5476 |
| Fuzhou | Wuxi | Shanghai | Beijing | Panyu | Guangzhou | Chongqi | Dazu | Mianning | Huangshan | |
|---|---|---|---|---|---|---|---|---|---|---|
| Fuzhou | ||||||||||
| Wuxi | 0.03516 ** | |||||||||
| Shanghai | 0.09036 ** | 0.06169 ** | ||||||||
| Beijing | 0.09087 ** | 0.08553 ** | 0.14506 ** | |||||||
| Panyu | 0.04102 ** | 0.01701 | 0.08110 ** | 0.06893 ** | ||||||
| Guangzhou | 0.05748 ** | 0.01548 | 0.09509 ** | 0.14693 ** | 0.02757 | |||||
| Chongqi | 0.06221 ** | 0.08104 ** | 0.15260** | 0.08600 * | 0.07274 ** | 0.13591 ** | ||||
| Dazu | 0.05353 ** | 0.02749 ** | 0.06415 ** | 0.06820 * | −0.00950 | 0.08728 * | 0.08683 ** | |||
| Mianning | 0.03928 ** | 0.05463 ** | 0.10666 ** | 0.11472 ** | 0.03229 * | 0.06161 ** | 0.08486 ** | 0.33040 | ||
| Huangshan | 0.02033 * | −0.00260 | 0.08273 ** | 0.09260 ** | 0.02300 | 0.03790 * | 0.05496 * | 0.03520 | 0.04779 ** | |
| Wenling | 0.02254 | −0.01230 | 0.06954 ** | 0.06412 * | −0.01610 | 0.00955 | 0.04418 | 0.01600 | 0.03280 * | −0.00860 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiu, Y.-f.; Liu, C.-C.; Xu, S.-h.; Lin, C.-S.; Chou, C.-C. The Genetic Diversity and Population Genetic Structure of the Red Panda, Ailurus fulgens, in Zoos in China. Animals 2020, 10, 1008. https://doi.org/10.3390/ani10061008
Xiu Y-f, Liu C-C, Xu S-h, Lin C-S, Chou C-C. The Genetic Diversity and Population Genetic Structure of the Red Panda, Ailurus fulgens, in Zoos in China. Animals. 2020; 10(6):1008. https://doi.org/10.3390/ani10061008
Chicago/Turabian StyleXiu, Yun-fang, Cheng-Chi Liu, Su-hui Xu, Chen-Si Lin, and Chin-Cheng Chou. 2020. "The Genetic Diversity and Population Genetic Structure of the Red Panda, Ailurus fulgens, in Zoos in China" Animals 10, no. 6: 1008. https://doi.org/10.3390/ani10061008
APA StyleXiu, Y.-f., Liu, C.-C., Xu, S.-h., Lin, C.-S., & Chou, C.-C. (2020). The Genetic Diversity and Population Genetic Structure of the Red Panda, Ailurus fulgens, in Zoos in China. Animals, 10(6), 1008. https://doi.org/10.3390/ani10061008





