Chemical Composition of Milk and Rumen Microbiome Diversity of Yak, Impacting by Herbage Grown at Different Phenological Periods on the Qinghai-Tibet Plateau
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Design
2.3. Sampling of Milk and Measurements of Chemical Composition
2.4. Sampling of Plant and Measurements of Chemical Composition
2.5. Sampling of Rumen Contents and Measurement of Fermentation Variables
2.6. Deoxyribonucleic Acid (DNA) Extraction, Amplification, and Sequencing
2.7. Sequencing Data Processing and Analysis
2.8. Statistical Analysis
3. Results
3.1. Nutritive Value of Yak Milk and Nutrient Compositions of Herbage
3.2. Rumen Volatile Fatty Acid Composition
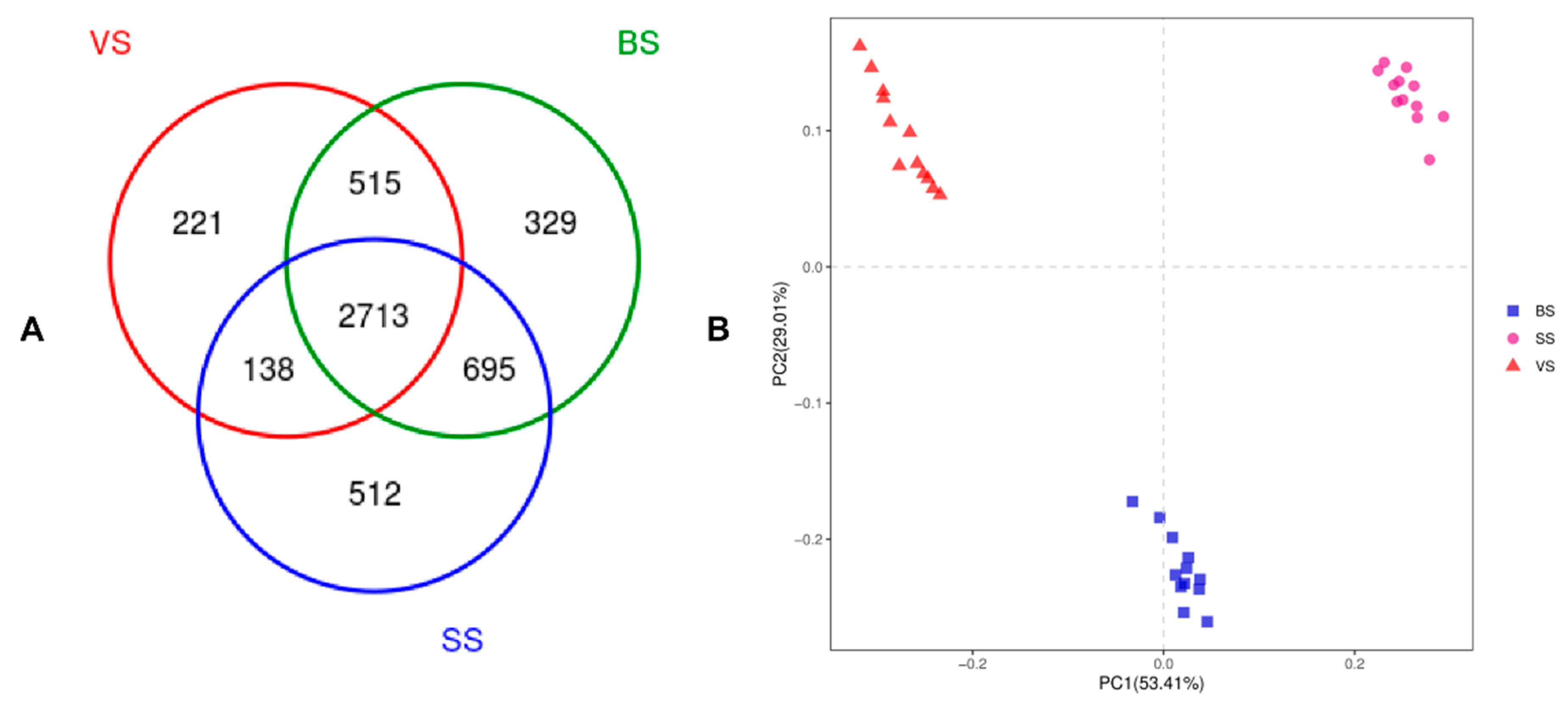
3.3. Bacterial Community Diversity, Richness and OTUs
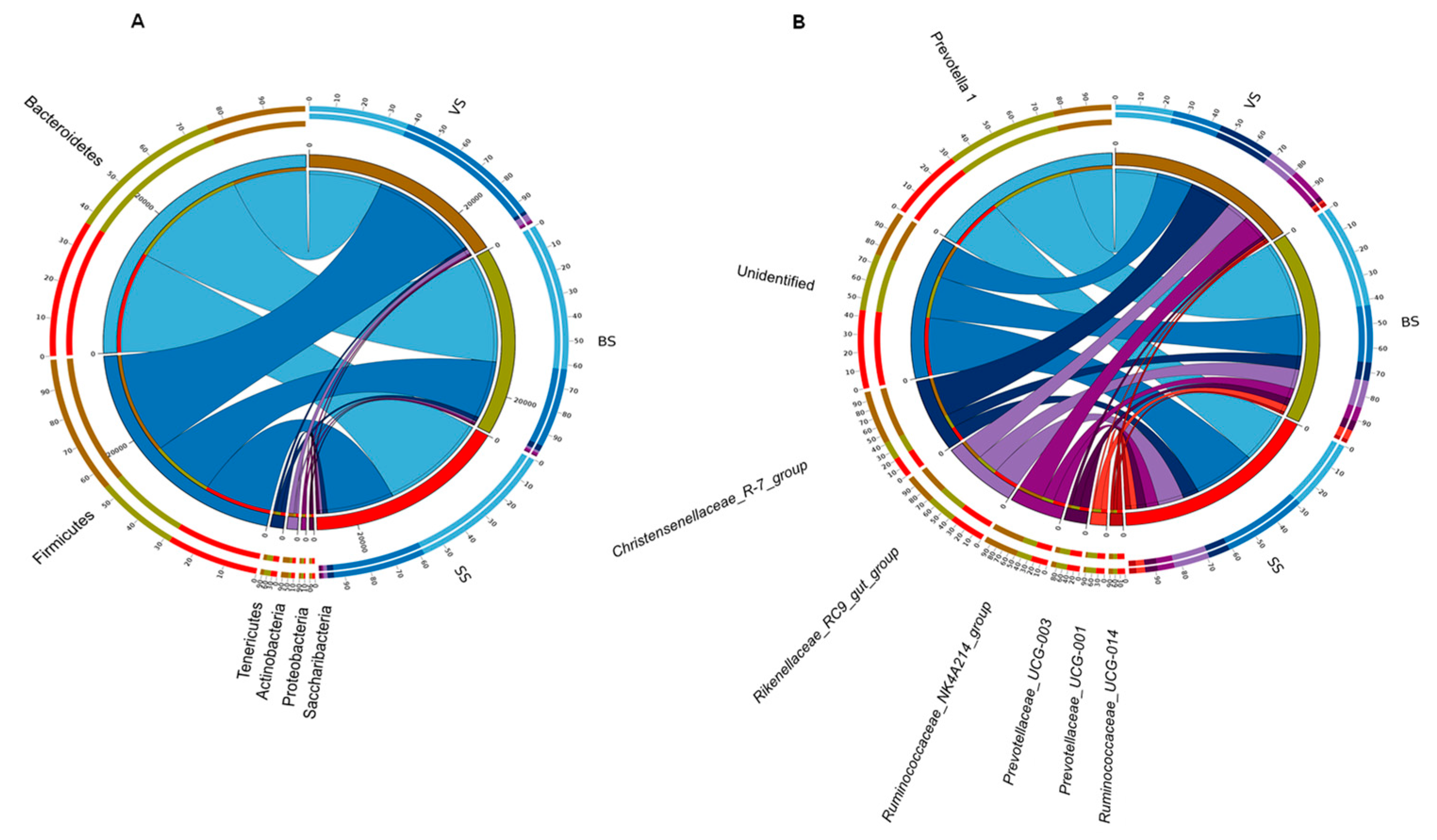
3.4. Rumen Bacterial Phylum and Genera
3.5. Milk Composition and Ruminal Fermentation Parameters in Relation to Nutrient Composition of Herbage
3.6. Milk Composition and Ruminal Fermentation Parameters in Relation to Main Bacteria at Genus Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Xia, W.; Osorio, J.S.; Yang, Y.; Liu, D.; Jiang, M.F. Short communication: Characterization of gene expression profiles related to yak milk protein synthesis during the lactation cycle. J. Dairy Sci. 2018, 101, 11150–11158. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Guo, Z.; Liu, Z.; Feng, Q.; Wang, X.; Tian, Q.; Ren, F.; Mao, X. The aggregation behavior and interactions of yak milk protein under thermal treatment. J. Dairy Sci. 2016, 99, 6137–6143. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Luo, Y.; Wang, D.H.; Kothapalli, K.S.; Brenna, T. Branched chain fatty acid composition of yak milk and manure during full-lactation and half-lactation. Prostaglandins Leukot. Essent. Fat. Acids 2019, 150, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Jiang, Y.; Chen, Y.; Zhao, Z.; Zhou, H.; Luo, Y.; Hu, J.; Hickford, J. Variation in the Fatty Acid Synthase Gene (FASN) and Its Association with Milk Traits in Gannan Yaks. Animals 2019, 9, 613. [Google Scholar] [CrossRef] [Green Version]
- Cui, G.; Yuan, F.; Degen, A.A.; Liu, S.; Zhou, J.; Shang, Z.; Ding, L.; Mi, J.; Wei, X.; Long, R. Composition of the milk of yaks raised at different altitudes on the Qinghai—Tibetan Plateau. Int. Dairy J. 2016, 59, 29–35. [Google Scholar] [CrossRef]
- Fan, Q.; Wanapat, M.; Hou, F. Mineral Nutritional Status of Yaks (Bos Grunniens) Grazing on the Qinghai-Tibetan Plateau. Animals 2019, 9, 468. [Google Scholar] [CrossRef] [Green Version]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F.; et al. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Fang, L.; Meng, Q.; Li, S.; Chai, S.; Liu, S.; Schonewille, J.T. Assessment of Ruminal Bacterial and Archaeal Community Structure in Yak (Bos grunniens). Front. Microbiol. 2017, 8, 207. [Google Scholar] [CrossRef] [Green Version]
- Henderson, G.; Global Rumen Census Collaborators; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- Bi, Y.; Zeng, S.; Zhang, R.; Diao, Q.; Tu, Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018, 18, 69. [Google Scholar] [CrossRef]
- Guo, W.; Bi, S.; Kang, J.; Zhang, Y.; Long, R.; Huang, X.; Shan, M.; Anderson, R.C. Bacterial communities related to 3-nitro-1-propionic acid degradation in the rumen of grazing ruminants in the Qinghai-Tibetan Plateau. Anaerobe 2018, 54, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Chen, H.; Chen, F.; He, Y.; Zhao, C.; Zhu, D.; Zeng, L.; Li, W. Analysis of the rumen bacteria and methanogenic archaea of yak (Bos grunniens) steers grazing on the Qinghai-Tibetan Plateau. Livest. Sci. 2016, 188, 61–71. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of the Association of Official Analytical Chemists International, 17th ed.; AOAC International: Gaithersburg, MD, USA, 2002. [Google Scholar]
- National Food Safety Standard of China. Determination of Lactose and Sucrose in Foods for Infants and Young Children Milk and Milk Products: GB 5413.21-2010; Ministry of Health of China: Beijing, China, 2010.
- Van Soest, P.; Robertson, J.; Lewis, B. Methods for Dietary Fiber, Neutral Detergent Fiber, and Nonstarch Polysaccharides in Relation to Animal Nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Shen, J.; Chai, Z.; Song, L.; Liu, J.; Wu, Y. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J. Dairy Sci. 2012, 95, 5978–5984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos-Morales, E.; Arco-Pérez, A.; Martín-García, A.; Yáñez-Ruiz, D.R.; Frutos, P.; Hervás, G. Use of stomach tubing as an alternative to rumen cannulation to study ruminal fermentation and microbiota in sheep and goats. Anim. Feed Sci. Technol. 2014, 198, 57–66. [Google Scholar] [CrossRef] [Green Version]
- Chaney, A.L.; Marbach, E.P. Modified Reagents for Determination of Urea and Ammonia. Clin. Chem. 1962, 8, 130–132. [Google Scholar] [CrossRef]
- Yu, Z.; Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 2004, 36, 808–812. [Google Scholar] [CrossRef]
- Dennis, K.L.; Wang, Y.; Blatner, N.R.; Wang, S.; Saadalla, A.; Trudeau, E.; Roers, A.; Weaver, C.T.; Lee, J.J.; Gilbert, J.A.; et al. Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by IL-10-producing T cells. Cancer Res. 2013, 73, 5905–5913. [Google Scholar] [CrossRef] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F. QIIME allows integration and analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yıldırım, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar] [CrossRef]
- Buxton, D.R. Quality-related characteristics of forages as influenced by plant environment and agronomic factors. Anim. Feed Sci. Technol. 1996, 59, 37–49. [Google Scholar] [CrossRef]
- Li, H.; Ma, Y.; Li, Q.; Wang, J.; Cheng, J.; Xue, S.J.; Shi, J. The Chemical Composition and Nitrogen Distribution of Chinese Yak (Maiwa) Milk. Int. J. Mol. Sci. 2011, 12, 4885–4895. [Google Scholar] [CrossRef]
- Liu, H.; Ren, F.; Jiang, L.; Ma, Z.; Qiao, H.; Zeng, S.; Gan, B.; Guo, H. Short communication: Fatty acid profile of yak milk from the Qinghai-Tibetan Plateau in different seasons and for different parities. J. Dairy Sci. 2011, 94, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Grummer, R.R. Effect of Feed on the Composition of Milk Fat. J. Dairy Sci. 1991, 74, 3244–3257. [Google Scholar] [CrossRef]
- Dijkstra, J.; Ellis, J.L.; Kebreab, E.; Strathe, A.; López, S.; France, J.; Bannink, A. Ruminal pH regulation and nutritional consequences of low pH. Anim. Feed Sci. Technol. 2012, 172, 22–33. [Google Scholar] [CrossRef]
- Weder, C.E.; DelCurto, T.; Svejcar, T.; Jaeger, J.; Bailey, R.K. Influence of supplemental alfalfa quality on the intake, use, and subsequent performance of beef cattle consuming low-quality roughages. J. Anim. Sci. 1999, 77, 1266–1276. [Google Scholar] [CrossRef] [Green Version]
- Nafikov, R.; Beitz, D. Carbohydrate and Lipid Metabolism in Farm Animals. J. Nutr. 2007, 137, 702–705. [Google Scholar] [CrossRef] [Green Version]
- Decuypere, J.A.; Dierick, N.A. The combined use of triacylglycerols containing medium-chain fatty acids and exogenous lipolytic enzymes as an alternative to in-feed antibiotics in piglets: Concept, possibilities and limitations. An overview. Nutr. Res. Rev. 2003, 16, 193–210. [Google Scholar] [CrossRef]
- Russell, J.B.; O’Connor, J.D.; Fox, D.G.; Van Soest, P.J.; Sniffen, C.J. A net carbohydrate and protein system for evaluating cattle diets: I. Ruminal fermentation. J. Anim. Sci. 1992, 70, 3551–3561. [Google Scholar] [CrossRef] [PubMed]
- Grummer, R.; Clark, J.; Davis, C.; Murphy, M. Effect of Ruminal Ammonia-Nitrogen Concentration on Protein Degradation in Situ. J. Dairy Sci. 1984, 67, 2294–2301. [Google Scholar] [CrossRef]
- Lv, X.; Chai, J.; Diao, Q.; Huang, W.; Zhuang, Y.; Zhang, N. The Signature Microbiota Drive Rumen Function Shifts in Goat Kids Introduced to Solid Diet Regimes. Microorganisms 2019, 7, 516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, F.; Wang, F.; King, M.; Wang, L. Effect of dynamic and static loading during in vitro degradation of a braided composite bioresorbable cardiovascular stent. Mater. Lett. 2019, 250, 12–15. [Google Scholar] [CrossRef]
- Suarez-Mena, F.; Heinrichs, A.; Jones, C.; Hill, T.; Quigley, J. Straw particle size in calf starters: Effects on digestive system development and rumen fermentation. J. Dairy Sci. 2015, 99, 341–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Lu, Z.; Xu, Z.; Chen, Z.; Shen, Z. Associations among dietary non-fiber carbohydrate, ruminal microbiota and epithelium G-protein-coupled receptor, and histone deacetylase regulations in goats. Microbiome 2017, 5, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkstra, J. Production and absorption of volatile fatty acids in the rumen. Livest. Prod. Sci. 1994, 39, 61–69. [Google Scholar] [CrossRef]
- Evans, N.J.; Brown, J.M.; Murray, R.D.; Getty, B.; Birtles, R.J.; Hart, C.A.; Carter, S. Characterization of Novel Bovine Gastrointestinal Tract Treponema Isolates and Comparison with Bovine Digital Dermatitis Treponemes. Appl. Environ. Microbiol. 2010, 77, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Goad, D.W.; Nagaraja, T.G. Ruminal microbial and fermentative changes associated with experimentally induced subacute acidosis in steers. J. Anim. Sci. 1998, 76, 234. [Google Scholar] [CrossRef] [Green Version]
- Jolazadeh, A.; Dehghan-Banadaky, M.; Rezayazdi, K. Effects of soybean meal treated with tannins extracted from pistachio hulls on performance, ruminal fermentation, blood metabolites and nutrient digestion of Holstein bulls. Anim. Feed Sci. Technol. 2015, 203, 33–40. [Google Scholar] [CrossRef]
- Wetzels, S.; Mann, E.; Metzler-Zebeli, B.; Wagner, M.; Klevenhusen, F.; Zebeli, Q.; Schmitz-Esser, S. Pyrosequencing reveals shifts in the bacterial epimural community relative to dietary concentrate amount in goats. J. Dairy Sci. 2015, 98, 5572–5587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huws, S.A.; Kim, E.J.; Cameron, S.; Girdwood, S.E.; Davies, L.; Tweed, J.; Vallin, H.; Scollan, N.D. Characterization of the rumen lipidome and microbiome of steers fed a diet supplemented with flax and echium oil. Microb. Biotechnol. 2014, 8, 331–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajima, K.; Aminov, R.; Nagamine, T.; Matsui, H.; Nakamura, M.; Benno, Y. Diet-Dependent Shifts in the Bacterial Population of the Rumen Revealed with Real-Time PCR. Appl. Environ. Microbiol. 2001, 67, 2766–2774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernando, S.C.; Purvis, H.T.; Najar, F.Z.; Sukharnikov, L.O.; Krehbiel, C.R.; Nagaraja, T.G.; Roe, B.A.; DeSilva, U. Rumen Microbial Population Dynamics during Adaptation to a High-Grain Diet. Appl. Environ. Microbiol. 2010, 76, 7482–7490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pope, P.B.; Smith, W.; Denman, S.; Tringe, S.G.; Barry, K.; Hugenholtz, P.; McSweeney, C.; McHardy, A.C.; Morrison, M. Isolation of Succinivibrionaceae Implicated in Low Methane Emissions from Tammar Wallabies. Science 2011, 333, 646–648. [Google Scholar] [CrossRef]
- Li, R.W.; Wu, S.; Baldwin, R.L.; Li, W.; Li, C.-J. Perturbation Dynamics of the Rumen Microbiota in Response to Exogenous Butyrate. PLoS ONE 2012, 7, e29392. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Huang, C.; Wang, J.; Zhou, H.; Lu, Y.; Lou, L.; Zheng, J.; Tian, L.; Wang, X.; Cao, Z.; et al. Dysbiosis of intestinal microbiota and decrease in paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PLoS ONE 2017, 12, e0176583. [Google Scholar] [CrossRef]
- Purushe, J.; North American Consortium for Rumen Bacteria; Fouts, D.E.; Morrison, M.; White, B.A.; Mackie, R.I.; Coutinho, P.M.; Henrissat, B.; Nelson, W.C. Comparative Genome Analysis of Prevotella ruminicola and Prevotella bryantii: Insights into Their Environmental Niche. Microb. Ecol. 2010, 60, 721–729. [Google Scholar] [CrossRef]
- Thoetkiattikul, H.; Mhuantong, W.; Laothanachareon, T.; Tangphatsornruang, S.; Pattarajinda, V.; Eurwilaichitr, L.; Champreda, V. Comparative Analysis of Microbial Profiles in Cow Rumen Fed with Different Dietary Fiber by Tagged 16S rRNA Gene Pyrosequencing. Curr. Microbiol. 2013, 67, 130–137. [Google Scholar] [CrossRef]
- Emerson, E.L.; Weimer, P.J. Fermentation of model hemicelluloses by Prevotella strains and Butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Appl. Microbiol. Biotechnol. 2017, 101, 4269–4278. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Ogata, K.; Tajima, K.; Nakamura, M.; Nagamine, T.; Aminov, R.; Benno, Y. Phenotypic characterization of polysaccharidases produced by four Prevotella type strains. Curr. Microbiol. 2000, 41, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Ushida, K.; Miyazaki, K.; Kojima, Y. Use of ratio of digested xylan to digested cellulose (X/C) as an index of fiber digestion in plant cell-wall material by ruminal microorganisms. Anim. Feed Sci. Technol. 1998, 71, 207–215. [Google Scholar] [CrossRef]
- Chiquette, J.; Allison, M.; Rasmussen, M. Prevotella bryantii 25A Used as a Probiotic in Early-Lactation Dairy Cows: Effect on Ruminal Fermentation Characteristics, Milk Production, and Milk Composition. J. Dairy Sci. 2008, 91, 3536–3543. [Google Scholar] [CrossRef]
- Russell, J.B.; Hespell, R.B. Microbial Rumen Fermentation. J. Dairy Sci. 1981, 64, 1153–1169. [Google Scholar] [CrossRef]



| Phenological Periods 1 | Major Species Communities in the Grassland | Above-Ground Biomass/(g/m2) | High Quality Herbage/(g/m2) | Other Herbage/(g/m2) |
|---|---|---|---|---|
| VS | Elymus nutans, Poa pratensi, Kobresia graminifolia | 43.8 | 28.6 | 15.1 |
| BS | Elymus nutans, Poa pratensi, Kobresia humili, Potentill abifurca, Saussurea pulchra, Ajania tenuifolia | 286.5 | 163.2 | 123.3 |
| SS | Elymus nutans | 121.4 | 76.4 | 45.1 |
| Nutrient Levels 2 | Phenological Periods 1, % of DM | SEM 3 | p-Value | ||
|---|---|---|---|---|---|
| VS | BS | SS | |||
| DM | 34.8 c | 46.8 b | 64.5 a | 0.29 | 0.025 |
| CP | 8.8 b | 12.8 a | 6.7 c | 0.44 | <0.01 |
| EE | 1.3 b | 1.8 a | 1.3 b | 0.04 | <0.01 |
| OM | 89.1 | 89.3 | 88.5 | 0.21 | 0.356 |
| NDF | 54.1 b | 51.2 c | 62.3 a | 0.84 | <0.01 |
| ADF | 35.1 b | 32.2 c | 36.3 a | 0.36 | <0.01 |
| Component | Phenological Periods 1 | SEM 2 | p-Value | ||
|---|---|---|---|---|---|
| VS | BS | SS | |||
| Milk yield, kg/d | 1.5 b | 2.4 a | 1.1 c | 0.03 | <0.01 |
| Fat, g/100 g | 6.2 b | 5.4 b | 6.7 a | 0.10 | <0.01 |
| Protein, g/100 g | 5.7 a | 5.7 a | 4.4 b | 0.18 | <0.01 |
| Lactose, g/100 g | 5.5 b | 5.9 a | 4.8 c | 0.10 | 0.026 |
| Total solids, g/100 g | 17.7 a | 14.5 b | 17.8 a | 0.25 | 0.030 |
| Item | Phenological Periods 1 | SEM 2 | p-Value | ||
|---|---|---|---|---|---|
| VS | BS | SS | |||
| pH | 7.4 a | 7.3 b | 7.4 a | 0.04 | 0.028 |
| Ammonia N, mg/L | 58.4 b | 96.5 a | 61.4 b | 9.18 | <0.01 |
| Concentration, mmol/L | |||||
| Total VFA 3 | 47.3 a | 49.4 a | 38.6 b | 1.55 | <0.01 |
| Acetate | 36.2 a | 35.5 a | 29.5 b | 1.08 | 0.021 |
| Propionate | 5.3 b | 7.5 a | 4.3 c | 0.23 | <0.01 |
| Butyrate | 4.6 a | 4.6 a | 3.2 b | 0.16 | 0.003 |
| Isobutyrate | 0.5 b | 0.7 a | 0.3 c | 0.02 | <0.01 |
| Valerate | 0.3 b | 0.4 a | 0.3 b | 0.01 | <0.01 |
| Isovalerate | 0.8 a | 0.6 c | 0.7 b | 0.02 | <0.01 |
| Molar proportion, % | |||||
| Acetate | 76.3 a | 71.9 b | 76.6 a | 0.46 | 0.036 |
| Propionate | 11.5 b | 15.3 a | 11.3 b | 0.16 | 0.014 |
| Butyrate | 9.8 | 9.3 | 8.2 | 0.29 | 0.072 |
| Isobutyrate | 1.1 b | 1.4 a | 0.7 c | 0.02 | <0.01 |
| Valerate | 0.6 | 0.8 | 0.7 | 0.03 | 0.206 |
| Isovalerate | 1.6 b | 1.2 c | 1.8 a | 0.02 | <0.01 |
| Acetate/propionate | 6.9 a | 4.6 b | 6.9 a | 0.25 | 0.022 |
| Bacterial Taxa | Phenological Periods 2, % | SEM 3 | p-Value | ||
|---|---|---|---|---|---|
| VS | BS | SS | |||
| Phylum level | |||||
| Synergistetes | 1.35 b | 1.21 c | 1.48 a | 0.01 | <0.01 |
| Proteobacteria | 1.48 b | 1.74 a | 1.13 c | 0.04 | 0.009 |
| Actinobacteria | 2.42 a | 1.37 c | 1.72 b | 0.11 | <0.01 |
| Tenericutes | 1.46 b | 2.41 a | 2.68 a | 0.10 | <0.01 |
| Firmicutes | 43.25 b | 57.36 a | 46.53 b | 1.84 | 0.023 |
| Bacteroidetes | 48.36 a | 34.24 c | 43.73 b | 1.78 | 0.018 |
| Genus level | |||||
| Firmicutes | |||||
| Christensenellaceae_R-7_group | 4.56 b | 6.38 a | 3.84 b | 0.80 | 0.035 |
| Ruminococcaceae_NK4A214_group | 4.64 b | 7.52 a | 4.83 b | 0.39 | <0.01 |
| Ruminococcaceae_UCG-010 | 1.38 b | 1.24 b | 2.01 a | 0.07 | <0.01 |
| Butyrivibrio_2 | 1.63 b | 2.43 a | 1.28 c | 0.14 | 0.028 |
| Ruminococcaceae_UCG-005 | 1.28 b | 1.41 b | 2.32 a | 0.09 | <0.01 |
| Eubacterium_coprostanoligenes_group | 0.73 c | 1.25 b | 1.64 a | 0.07 | <0.01 |
| Ruminococcaceae_UCG-014 | 1.36 b | 2.26 a | 2.42 a | 0.06 | <0.01 |
| Succiniclasticum | 1.24 b | 2.21 a | 1.07 b | 0.08 | 0.005 |
| Actinobacteria | |||||
| Olsenella | 0.93 a | 0.76 b | 1.03 a | 0.05 | <0.01 |
| Senegalimassilia | 1.84 a | 1.31 b | 1.22 b | 0.03 | 0.026 |
| Proteobacteria | |||||
| Desulfovibrio | 0.58 b | 1.06 a | 0.14 c | 0.01 | <0.01 |
| Bacteroidetes | |||||
| Prevotella_1 | 19.42 b | 16.53 c | 23.65 a | 1.21 | 0.024 |
| Rikenellaceae_RC9_gut_group | 7.12 b | 6.72 b | 9.14 a | 0.16 | <0.01 |
| Prevotellaceae_UCG-003 | 1.57 c | 2.43 b | 3.33 a | 0.15 | <0.01 |
| Prevotellaceae_UCG-001 | 3.23 a | 1.84 c | 2.20 b | 0.15 | 0.022 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Q.; Wanapat, M.; Hou, F. Chemical Composition of Milk and Rumen Microbiome Diversity of Yak, Impacting by Herbage Grown at Different Phenological Periods on the Qinghai-Tibet Plateau. Animals 2020, 10, 1030. https://doi.org/10.3390/ani10061030
Fan Q, Wanapat M, Hou F. Chemical Composition of Milk and Rumen Microbiome Diversity of Yak, Impacting by Herbage Grown at Different Phenological Periods on the Qinghai-Tibet Plateau. Animals. 2020; 10(6):1030. https://doi.org/10.3390/ani10061030
Chicago/Turabian StyleFan, Qingshan, Metha Wanapat, and Fujiang Hou. 2020. "Chemical Composition of Milk and Rumen Microbiome Diversity of Yak, Impacting by Herbage Grown at Different Phenological Periods on the Qinghai-Tibet Plateau" Animals 10, no. 6: 1030. https://doi.org/10.3390/ani10061030
APA StyleFan, Q., Wanapat, M., & Hou, F. (2020). Chemical Composition of Milk and Rumen Microbiome Diversity of Yak, Impacting by Herbage Grown at Different Phenological Periods on the Qinghai-Tibet Plateau. Animals, 10(6), 1030. https://doi.org/10.3390/ani10061030






