Edition of Prostaglandin E2 Receptors EP2 and EP4 by CRISPR/Cas9 Technology in Equine Adipose Mesenchymal Stem Cells
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. CRISPR
2.2.1. CRISPR Design
2.2.2. Cloning and Hybridisation of gRNA Oligonucleotides
2.3. Lentivirus Production and Transfection
2.4. Transduction of aMSCs and Cell Clones
2.5. PCR and T7 Endonuclease I (T7EI) Cleavage Assays for Detection of Insertions and Deletions of Clones
2.6. Cloning and Analysis of Indel (Random Insertion or Deletion) Frequencies
2.7. RNA Isolation and Quantitative Real-Time Reverse Transcriptase PCR Analysis
2.8. Immunocytochemical Analyses
2.9. Cell Migration Assessed by Scratch Assay
2.10. Expression MSC Surface Markers
2.11. Statistical Analysis
3. Results
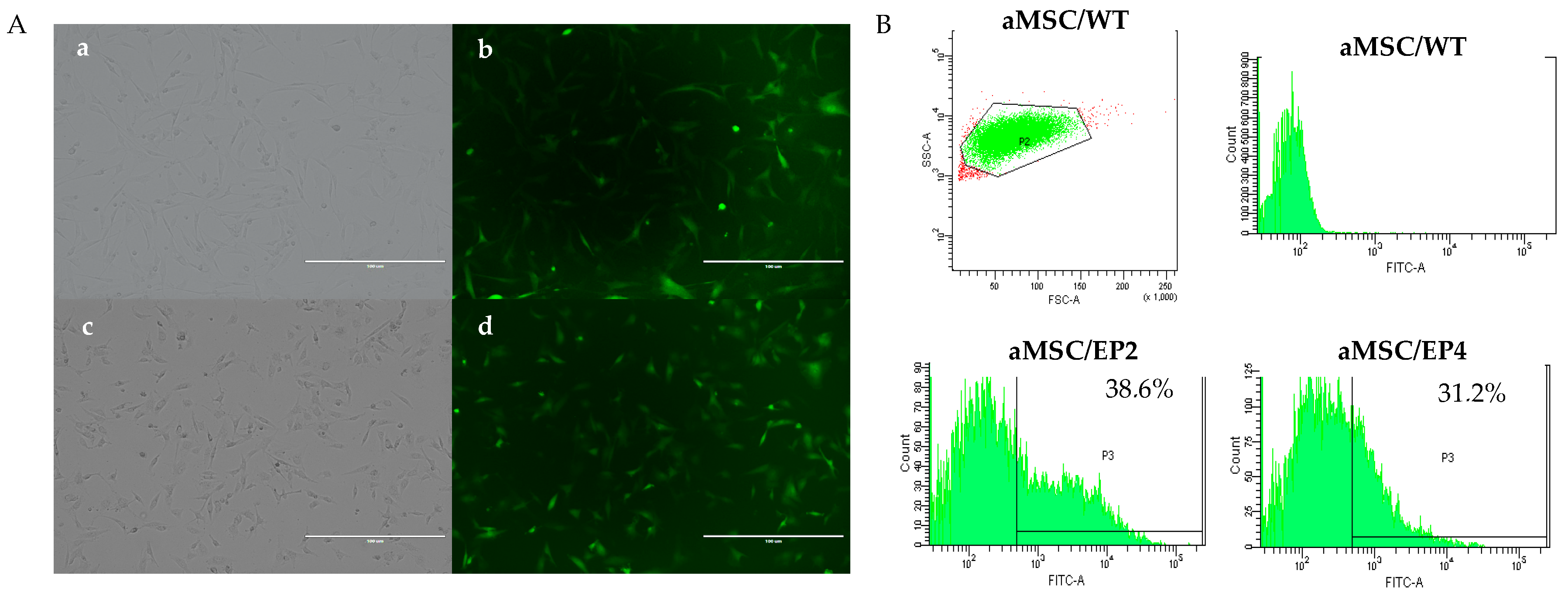
3.1. Transduction of aMSCs and Culture of Cellular Clones
3.2. T7EI Test, Cloning, and Frequencies of Indels
3.3. Immunocytochemistry and Gene Expression for EP2 and EP4 Receptors
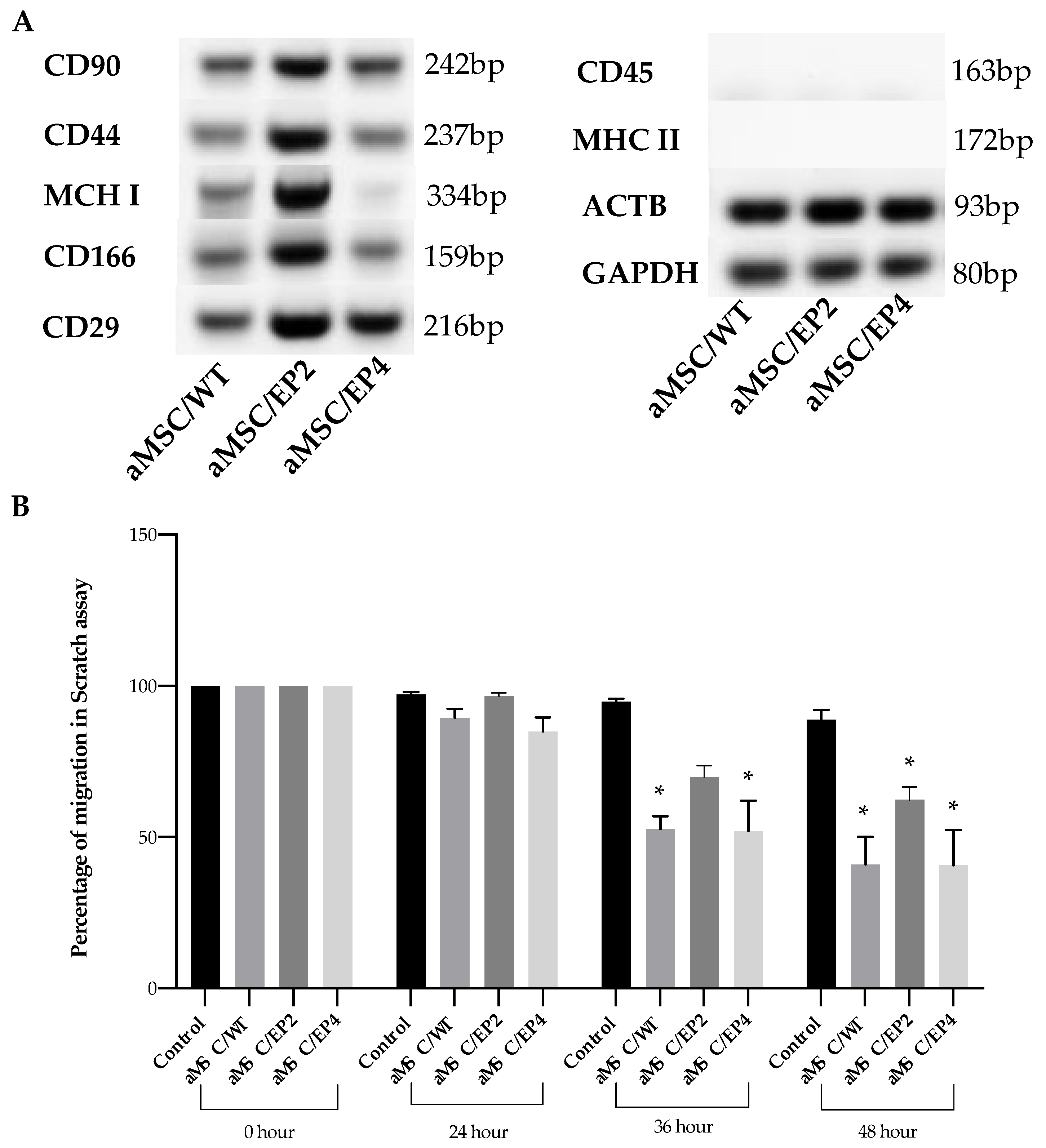
3.4. MSC Surface Markers and MSC Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lee, B.C.; Kim, H.S.; Shin, T.H.; Kang, I.; Lee, J.Y.; Kim, J.J.; Kang, H.K.; Seo, Y.; Lee, S.; Yu, K.R.; et al. PGE2 maintains self-renewal of human adult stem cells via EP2-mediated autocrine signaling and its production is regulated by cell-to-cell contact. Sci. Rep. 2016, 6, 26298. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Narumiya, S. Prostaglandin E receptors. J. Biol. Chem. 2007, 282, 11613–11617. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, I.M.; Price, S.B.; Cronin, J.; Gilbert, R.O.; Gadsby, J.E. Mechanisms of infertility associated with clinical and subclinical endometritis in high producing dairy cattle. Reprod. Domest. Anim. 2009, 44 (Suppl. 3), 1–9. [Google Scholar] [CrossRef]
- Regan, J.W. EP2 and EP4 prostanoid receptor signaling. Life Sci. 2003, 74, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.L.; DeWitt, D.L.; Garavito, R.M. Cyclooxygenases: Structural, cellular, and molecular biology. Annu. Rev. Biochem. 2000, 69, 145–182. [Google Scholar] [CrossRef] [PubMed]
- Vukicevic, S.; Simic, P.; Borovecki, F.; Grgurevic, L.; Rogic, D.; Orlic, I.; Grasser, W.A.; Thompson, D.D.; Paralkar, V.M. Role of EP2 and EP4 receptor-selective agonists of prostaglandin E(2) in acute and chronic kidney failure. Kidney. Int. 2006, 70, 1099–1106. [Google Scholar] [CrossRef]
- Ushikubi, F.; Sugimoto, Y.; Ichikawa, A.; Narumiya, S. Roles of prostanoids revealed from studies using mice lacking specific prostanoid receptors. JPN J. Pharm. 2000, 83, 279–285. [Google Scholar] [CrossRef]
- Wånggren, K.; Lalitkumar, P.G.; Stavreus-Evers, A.; Ståbi, B.; Gemzell-Danielsson, K. Prostaglandin E2 and F2alpha receptors in the human Fallopian tube before and after mifepristone treatment. Mol. Hum. Reprod. 2006, 12, 577–585. [Google Scholar] [CrossRef]
- Breyer, R.M.; Bagdassarian, C.K.; Myers, S.A.; Breyer, M.D. Prostanoid receptors: Subtypes and signaling. Annu. Rev. Pharm. Toxicol. 2001, 41, 661–690. [Google Scholar] [CrossRef]
- Wang, Y.; Lai, S.; Tang, J.; Feng, C.; Liu, F.; Su, C.; Zou, W.; Chen, H.; Xu, D. Prostaglandin E2 promotes human CD34+ cells homing through EP2 and EP4 in vitro. Mol. Med. Rep. 2017, 16, 639–646. [Google Scholar] [CrossRef]
- Samuelsson, B.; Morgenstern, R.; Jakobsson, P.J. Membrane prostaglandin E synthase-1: A novel therapeutic target. Pharm. Rev. 2007, 59, 207–224. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lu, X.; Zou, L.; Xu, X.; Qiu, H. E-Prostanoid 2 Receptor Overexpression Promotes Mesenchymal Stem Cell Attenuated Lung Injury. Hum. Gene. Ther. 2016, 27, 621–630. [Google Scholar] [CrossRef]
- Lu, X.; Han, J.; Xu, X.; Xu, J.; Liu, L.; Huang, Y.; Yang, Y.; Qiu, H. PGE2 Promotes the Migration of Mesenchymal Stem Cells through the Activation of FAK and ERK1/2 Pathway. Stem. Cells Int. 2017, 2017, 8178643. [Google Scholar] [CrossRef] [PubMed]
- Hoggatt, J.; Singh, P.; Sampath, J.; Pelus, L.M. Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood 2009, 113, 5444–5455. [Google Scholar] [CrossRef] [PubMed]
- Carrade, D.D.; Lame, M.W.; Kent, M.S.; Clark, K.C.; Walker, N.J.; Borjesson, D.L. Comparative Analysis of the Immunomodulatory Properties of Equine Adult-Derived Mesenchymal Stem Cells. Cell Med. 2012, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.M.; Pindjakova, J.; Hanley, S.A.; McCarthy, C.; Weidhofer, G.A.; Sweeney, E.M.; English, K.; Shaw, G.; Murphy, J.M.; Barry, F.P.; et al. Mesenchymal stem cell inhibition of T-helper 17 cell- differentiation is triggered by cell-cell contact and mediated by prostaglandin E2 via the EP4 receptor. Eur. J. Immunol. 2011, 41, 2840–2851. [Google Scholar] [CrossRef]
- Matysiak, M.; Orlowski, W.; Fortak-Michalska, M.; Jurewicz, A.; Selmaj, K. Immunoregulatory function of bone marrow mesenchymal stem cells in EAE depends on their differentiation state and secretion of PGE2. J. Neuroimmunol. 2011, 233, 106–111. [Google Scholar] [CrossRef]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef]
- Niu, J.; Zhang, B.; Chen, H. Applications of TALENs and CRISPR/Cas9 in human cells and their potentials for gene therapy. Mol. Biotechnol. 2014, 56, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, J.; Yu, T.; Qi, C. Generation of PTEN knockout bone marrow mesenchymal stem cell lines by CRISPR/Cas9-mediated genome editing. Cytotechnology 2018, 70, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Malik, A.A.; Ahmad, S.M.; Shah, R.A.; Ganai, N.A.; Shafi, S.S.; Shabir, N. Advances in genome editing for improved animal breeding: A review. Vet. World 2017, 10, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Zarei, A.; Razban, V.; Hosseini, S.E.; Tabei, S.M.B. Creating cell and animal models of human disease by genome editing using CRISPR/Cas9. J. Gene Med. 2019, 21, e3082. [Google Scholar] [CrossRef] [PubMed]
- Cabezas, J.; Rojas, D.; Navarrete, F.; Ortiz, R.; Rivera, G.; Saravia, F.; Rodriguez-Alvarez, L.; Castro, F.O. Equine mesenchymal stem cells derived from endometrial or adipose tissue share significant biological properties, but have distinctive pattern of surface markers and migration. Theriogenology 2018, 106, 93–102. [Google Scholar] [CrossRef]
- Castro, F.O.; Torres, A.; Cabezas, J.; Rodríguez-Alvarez, L. Combined use of platelet rich plasma and vitamin C positively affects differentiation in vitro to mesodermal lineage of adult adipose equine mesenchymal stem cells. Res. Vet. Sci. 2014, 96, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Bundgaard, L.; Stensballe, A.; Elbæk, K.J.; Berg, L.C. Mapping of equine mesenchymal stromal cell surface proteomes for identification of specific markers using proteomics and gene expression analysis: An in vitro cross-sectional study. Stem. Cell Res. Ther. 2018, 9, 288. [Google Scholar] [CrossRef] [PubMed]
- Connick, P.; Kolappan, M.; Crawley, C.; Webber, D.J.; Patani, R.; Michell, A.W.; Du, M.-Q.; Luan, S.-L.; Altmann, D.R.; Thompson, A.J.; et al. Autologous mesenchymal stem cells for the treatment of secondary progressive multiple sclerosis: An open-label phase 2a proof-of-concept study. Lancet Neurol. 2012, 11, 150–156. [Google Scholar] [CrossRef]
- Hmadcha, A.; Martin-Montalvo, A.; Gauthier, B.R.; Soria, B.; Capilla-Gonzalez, V. Therapeutic Potential of Mesenchymal Stem Cells for Cancer Therapy. Front. Bioeng. Biotechnol. 2020, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Pittenger, M.F. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood 2005, 105, 1815–1822. [Google Scholar] [CrossRef]
- Gao, F.; Chiu, S.M.; Motan, D.A.; Zhang, Z.; Chen, L.; Ji, H.L.; Tse, H.F.; Fu, Q.L.; Lian, Q. Mesenchymal stem cells and immunomodulation: Current status and future prospects. Cell Death Dis. 2016, 7, e2062. [Google Scholar] [CrossRef] [PubMed]
- Food, U.; Administration, D. Cellular products for joint surface repair. Cell. Tissue Gene Ther. Advis. Comm. 2005. [Google Scholar]
- Paris, D.B.; Stout, T.A. Equine embryos and embryonic stem cells: Defining reliable markers of pluripotency. Theriogenology 2010, 74, 516–524. [Google Scholar] [CrossRef]
- Mambelli, L.I.; Winter, G.H.; Kerkis, A.; Malschitzky, E.; Mattos, R.C.; Kerkis, I. A novel strategy of mesenchymal stem cells delivery in the uterus of mares with endometrosis. Theriogenology 2013, 79, 744–750. [Google Scholar] [CrossRef]
- Rink, B.E.; Beyer, T.; French, H.M.; Watson, E.; Aurich, C.; Donadeu, F.X. The Fate of Autologous Endometrial Mesenchymal Stromal Cells After Application in the Healthy Equine Uterus. Stem. Cells Dev. 2018, 27, 1046–1052. [Google Scholar] [CrossRef]
- Jang, M.W.; Yun, S.P.; Park, J.H.; Ryu, J.M.; Lee, J.H.; Han, H.J. Cooperation of Epac1/Rap1/Akt and PKA in prostaglandin E(2)-induced proliferation of human umbilical cord blood derived mesenchymal stem cells: Involvement of c-Myc and VEGF expression. J. Cell Physiol. 2012, 227, 3756–3767. [Google Scholar] [CrossRef]
- Kleiveland, C.R.; Kassem, M.; Lea, T. Human mesenchymal stem cell proliferation is regulated by PGE2 through differential activation of cAMP-dependent protein kinase isoforms. Exp. Cell Res. 2008, 314, 1831–1838. [Google Scholar] [CrossRef]
- Osma-Garcia, I.C.; Punzón, C.; Fresno, M.; Díaz-Muñoz, M.D. Dose-dependent effects of prostaglandin E2 in macrophage adhesion and migration. Eur. J. Immunol. 2016, 46, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Concordet, J.P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucl. Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Petersen, G.F.; Hilbert, B.; Trope, G.; Kalle, W.; Strappe, P. Efficient transduction of equine adipose-derived mesenchymal stem cells by VSV-G pseudotyped lentiviral vectors. Res. Vet. Sci. 2014, 97, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Kallifatidis, G.; Beckermann, B.M.; Groth, A.; Schubert, M.; Apel, A.; Khamidjanov, A.; Ryschich, E.; Wenger, T.; Wagner, W.; Diehlmann, A.; et al. Improved lentiviral transduction of human mesenchymal stem cells for therapeutic intervention in pancreatic cancer. Cancer Gene Ther. 2008, 15, 231–240. [Google Scholar] [CrossRef]
- Van Damme, A.; Thorrez, L.; Ma, L.; Vandenburgh, H.; Eyckmans, J.; Dell’Accio, F.; De Bari, C.; Luyten, F.; Lillicrap, D.; Collen, D.; et al. Efficient Lentiviral Transduction and Improved Engraftment of Human Bone Marrow Mesenchymal Cells. Stem. Cells 2006, 24, 896–907. [Google Scholar] [CrossRef]
- Chen, L.; Qu, J.; Cheng, T.; Chen, X.; Xiang, C. Menstrual blood-derived stem cells: Toward therapeutic mechanisms, novel strategies, and future perspectives in the treatment of diseases. Stem. Cell Res. Ther. 2019, 10, 406. [Google Scholar] [CrossRef]
- Goujon, C.; Jarrosson-Wuillème, L.; Bernaud, J.; Rigal, D.; Darlix, J.L.; Cimarelli, A. With a little help from a friend: Increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC). Gene 2006, 13, 991–994. [Google Scholar] [CrossRef]
- Lin, P.; Lin, Y.; Lennon, D.P.; Correa, D.; Schluchter, M.; Caplan, A.I. Efficient lentiviral transduction of human mesenchymal stem cells that preserves proliferation and differentiation capabilities. Stem. Cells Transl. Med. 2012, 1, 886–897. [Google Scholar] [CrossRef]
- De Oliveira, V.C.; Moreira, G.S.A.; Bressan, F.F.; Gomes Mariano Junior, C.; Roballo, K.C.S.; Charpentier, M.; Concordet, J.P.; Meirelles, F.V.; Ambrósio, C.E. Edition of TFAM gene by CRISPR/Cas9 technology in bovine model. PLoS ONE 2019, 14, e0213376. [Google Scholar] [CrossRef]
- Sentmanat, M.F.; Peters, S.T.; Florian, C.P.; Connelly, J.P.; Pruett-Miller, S.M. A Survey of Validation Strategies for CRISPR-Cas9 Editing. Sci. Rep. 2018, 8, 888. [Google Scholar] [CrossRef]
- Jia, C.; Huai, C.; Ding, J.; Hu, L.; Su, B.; Chen, H.; Lu, D. New applications of CRISPR/Cas9 system on mutant DNA detection. Gene 2018, 641, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Hsiau, T.; Conant, D.; Rossi, N.; Maures, T.; Waite, K.; Yang, J.; Joshi, S.; Kelso, R.; Holden, K.; Enzmann, B.L.; et al. Inference of CRISPR Edits from Sanger Trace Data. BioRxiv 2019. [Google Scholar] [CrossRef]
- Jin, J.; Xu, Y.; Huo, L.; Ma, L.; Scott, A.W.; Pizzi, M.P.; Li, Y.; Wang, Y.; Yao, X.; Song, S.; et al. An improved strategy for CRISPR/Cas9 gene knockout and subsequent wildtype and mutant gene rescue. PLoS ONE 2020, 15, e0228910. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Alander, C.; Zhan, P.; Gao, Q.; Pilbeam, C.; Raisz, L. Effect of deletion of the prostaglandin EP2 receptor on the anabolic response to prostaglandin E2 and a selective EP2 receptor agonist. Prostaglandins Other Lipid Mediat. 2008, 86, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Ball, B.A.; Scoggin, K.E.; Troedsson, M.H.; Squires, E.L. Characterization of prostaglandin E2 receptors (EP2, EP4) in the horse oviduct. Anim. Reprod. Sci. 2013, 142, 35–41. [Google Scholar] [CrossRef]
- Delville, M.; Soheili, T.; Bellier, F.; Durand, A.; Denis, A.; Lagresle-Peyrou, C.; Cavazzana, M.; Andre-Schmutz, I.; Six, E. A Nontoxic Transduction Enhancer Enables Highly Efficient Lentiviral Transduction of Primary Murine T Cells and Hematopoietic Stem Cells. Mol. Ther. Method. Clin. Dev. 2018, 10, 341–347. [Google Scholar] [CrossRef]
- Schomann, T.; Mezzanotte, L.; Lourens, I.M.; de Groot, J.C.; Frijns, J.H.; Huisman, M.A. Lentiviral transduction and subsequent loading with nanoparticles do not affect cell viability and proliferation in hair-follicle-bulge-derived stem cells in vitro. Contrast Media Mol. Imaging 2016, 11, 550–560. [Google Scholar] [CrossRef]
- Hizaki, H.; Segi, E.; Sugimoto, Y.; Hirose, M.; Saji, T.; Ushikubi, F.; Matsuoka, T.; Noda, Y.; Tanaka, T.; Yoshida, N.; et al. Abortive expansion of the cumulus and impaired fertility in mice lacking the prostaglandin E receptor subtype EP(2). Proc. Natl. Acad. Sci. USA 1999, 96, 10501–10506. [Google Scholar] [CrossRef]
- Tilley, S.L.; Audoly, L.P.; Hicks, E.H.; Kim, H.S.; Flannery, P.J.; Coffman, T.M.; Koller, B.H. Reproductive failure and reduced blood pressure in mice lacking the EP2 prostaglandin E2 receptor. J. Clin. Investig. 1999, 103, 1539–1545. [Google Scholar] [CrossRef]
- Biswas, S.; Bhattacherjee, P.; Paterson, C.A.; Tilley, S.L.; Koller, B.H. Ocular inflammatory responses in the EP2 and EP4 receptor knockout mice. Ocul. Immunol. Inflamm. 2006, 14, 157–163. [Google Scholar] [CrossRef]
- Yun, S.P.; Ryu, J.M.; Jang, M.W.; Han, H.J. Interaction of profilin-1 and F-actin via a β-arrestin-1/JNK signaling pathway involved in prostaglandin E(2)-induced human mesenchymal stem cells migration and proliferation. J. Cell. Physiol. 2011, 226, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.A.; Bhagat, S.; Alam, M.I. PGE(2) -induced migration of human brain endothelial cell is mediated though protein kinase A in cooperation of EP receptors. J. Leukoc. Biol. 2019, 105, 705–717. [Google Scholar] [CrossRef] [PubMed]
- De Schauwer, C.; Meyer, E.; Van de Walle, G.R.; Van Soom, A. Markers of stemness in equine mesenchymal stem cells: A plea for uniformity. Theriogenology 2011, 75, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Barberini, D.J.; Freitas, N.P.; Magnoni, M.S.; Maia, L.; Listoni, A.J.; Heckler, M.C.; Sudano, M.J.; Golim, M.A.; da Cruz Landim-Alvarenga, F.; Amorim, R.M. Equine mesenchymal stem cells from bone marrow, adipose tissue and umbilical cord: Immunophenotypic characterization and differentiation potential. Stem Cell Res. Ther. 2014, 5, 25. [Google Scholar] [CrossRef]
- De Schauwer, C.; Piepers, S.; Van de Walle, G.R.; Demeyere, K.; Hoogewijs, M.K.; Govaere, J.L.; Braeckmans, K.; Van Soom, A.; Meyer, E. In search for cross-reactivity to immunophenotype equine mesenchymal stromal cells by multicolor flow cytometry. Cytometry A 2012, 81, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Radcliffe, C.H.; Flaminio, M.J.; Fortier, L.A. Temporal analysis of equine bone marrow aspirate during establishment of putative mesenchymal progenitor cell populations. Stem Cells Dev. 2010, 19, 269–282. [Google Scholar] [CrossRef] [PubMed]



| Name | Sequence (5’–3’) | Product Size | Position Sequence |
|---|---|---|---|
| PTGER2_sgRNA | Fwd: CACCGTGGTGCTGGCTTCGTACGCG | 226/Fwd | |
| Rev: AAACCGCGTACGAAGCCAGCACCAC | |||
| PTGER2_Ontarget | Fwd: GGAGGACTCGTCTCCCTCTT | 393 bp | |
| Rev: AGGCTGAAGAAGGTCATGGC | |||
| PTGER4_sgRNA | FwdCACCGGAGACGACCTTCTACACGT | 230/Fwd | |
| Rev: AAACACGTGTAGAAGGTCGTCTCC | |||
| PTGER4_Ontarget | Fwd: GGACACGTAGACGGCAAAGA | 358 bp | |
| Rev: TGTTCATCTTCGGGGTGGTG |
| Acess number | Gene Symbol | Primer Sequence (5’-3’) | Product Size |
|---|---|---|---|
| NM_001127352.1 | PTGER2 | Fw: CATCAGCTCCGTGATGGTCT | 294 bp |
| Rev: ATCGTGGCCAGGCTGAAGA | |||
| XM_001499068.5 | PTGER4 | Fw: AGCTCCAACCTGCCCAAGAGT | 406 bp |
| Rev: CATTGGACACGTAGACGGCAAA | |||
| NM_001081838.1 | ACTB | Fw: GCTCCCAGCACGATGAAGAT | 93 bp |
| Rev: GGTGGACAATGAGGCCAGAA | |||
| NM_001163856.1 | GAPDH | Fw: GGGTGGAGCCAAAAGGGTCATCAT | 80 bp |
| Rev: AGCTTTCTCCAGGCGGCAGGTCAG | |||
| NM_001301217.1 | CD29 | Fw: GTGAGATGTGTCAGACGTGC | 216 bp |
| Rev: AGAACCAGCAGTCATCCACA | |||
| NM_001085435.2 | CD44 | Fw: TTCATAGAAGGGCACGTGGT | 237 bp |
| Rev: GCCTTTCTTGGTGTAGCGAG | |||
| XM_001503225.4 | CD90 | Fw: TCTCCTGCTGACAGTCTTGC | 242 bp |
| Rev: GGACCTTGATGTTGTACTTGC | |||
| XM_001503380.6 | CD166 | Fw: GCAGAAAACCAGCTGGAGAG | 159 bp |
| Rev: AGCGAGGAGTAGACCAACGA | |||
| NM_001309162.1 | CD45 | Fw: CTCCTCATTCACTGCAGAGA | 163 bp |
| Rev: GGTACTGCTCAAATGTGGGA | |||
| NM_001082508.2 | MHC I | Fw: TTCATCTCCGTCGGCTACGTG | 334 bp |
| Rev: AGGAGCGCAGGTCCTCGTT | |||
| NM_001142816.1 | MHC II | Fw: AGCGGCGAGTTGAACCTACAGT | 172 bp |
| Rev: CGGATCAGACCTGTGGAGATGA | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mançanares, A.C.F.; Cabezas, J.; Manríquez, J.; de Oliveira, V.C.; Wong Alvaro, Y.S.; Rojas, D.; Navarrete Aguirre, F.; Rodriguez-Alvarez, L.; Castro, F.O. Edition of Prostaglandin E2 Receptors EP2 and EP4 by CRISPR/Cas9 Technology in Equine Adipose Mesenchymal Stem Cells. Animals 2020, 10, 1078. https://doi.org/10.3390/ani10061078
Mançanares ACF, Cabezas J, Manríquez J, de Oliveira VC, Wong Alvaro YS, Rojas D, Navarrete Aguirre F, Rodriguez-Alvarez L, Castro FO. Edition of Prostaglandin E2 Receptors EP2 and EP4 by CRISPR/Cas9 Technology in Equine Adipose Mesenchymal Stem Cells. Animals. 2020; 10(6):1078. https://doi.org/10.3390/ani10061078
Chicago/Turabian StyleMançanares, Ana Carolina Furlanetto, Joel Cabezas, José Manríquez, Vanessa Cristina de Oliveira, Yat Sen Wong Alvaro, Daniela Rojas, Felipe Navarrete Aguirre, Lleretny Rodriguez-Alvarez, and Fidel Ovidio Castro. 2020. "Edition of Prostaglandin E2 Receptors EP2 and EP4 by CRISPR/Cas9 Technology in Equine Adipose Mesenchymal Stem Cells" Animals 10, no. 6: 1078. https://doi.org/10.3390/ani10061078
APA StyleMançanares, A. C. F., Cabezas, J., Manríquez, J., de Oliveira, V. C., Wong Alvaro, Y. S., Rojas, D., Navarrete Aguirre, F., Rodriguez-Alvarez, L., & Castro, F. O. (2020). Edition of Prostaglandin E2 Receptors EP2 and EP4 by CRISPR/Cas9 Technology in Equine Adipose Mesenchymal Stem Cells. Animals, 10(6), 1078. https://doi.org/10.3390/ani10061078






