Survey of Serum Amyloid A and Bacterial and Viral Frequency Using qPCR Levels in Recently Captured Feral Donkeys from Death Valley National Park (California)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
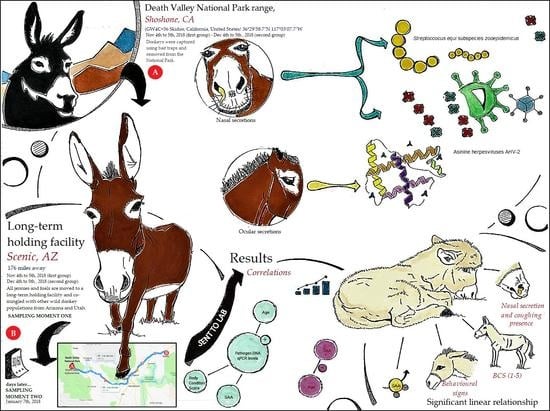
2.1. Animal Sample and Sampling
2.2. SAA Levels
2.3. Load Quantification for Viruses and Bacteria
2.4. Quantitative PCR Systems
2.5. RT-Reaction and Quantitative PCR
2.6. Statistical Analyses
2.7. Ethical Approval
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tiller, B. Feral Burro Populations: Distribution and Damage Assessment; Pacific Northwest National Laboratory: Richland, WA, USA, 1997. [Google Scholar]
- Weaver, R.A. Feral burros and wildlife. In Proceedings of the 6th Vertebrate Pest Conference, Anaheim, CA, USA, 5–7 March 1974. [Google Scholar]
- National Research Council. Using Science to Improve the BLM Wild Horse and Burro Program: A Way Forward; National Academies Press: Washington, DC, USA, 2013. [Google Scholar]
- National Research Council/Commission on Natural Resources/National Research Council; Committee on Wild, Free-Roaming Horses Burros. Wild and Free-Roaming Horses and Burros: Current Knowledge and Recommended Research; National Academies Press: Washington, DC, USA, 1980. [Google Scholar]
- Satué, K.; Calvo, A.; Gardón, J.C. Factors influencing serum amyloid type A (SAA) concentrations in horses. Open. J. Vet. Med. 2013, 3, 58. [Google Scholar] [CrossRef]
- Nolen-Walston, R. How to interpret serum amyloid A concentrations. In Proceedings of the 65th Annual American Association of Equine Practitioners (AAEP), Denver, CO, USA, 7–11 December 2019; pp. 130–137. [Google Scholar]
- Kay, G.; Tligui, N.; Semmate, N.; Azrib, R.; González, F.J.N.; Brizgys, L.; McLean, A. Determining factors and interspecific modeling for serum amyloid a concentrations in working horses, donkeys, and mules. Res. Vet. Sci. 2019, 125, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, S.; Andersen, P.H. The acute phase protein serum amyloid A (SAA) as a marker of inflammation in horses. Equine Vet. Educ. 2007, 19, 38–46. [Google Scholar] [CrossRef]
- Pusterla, N.; Kass, P.H.; Mapes, S.; Johnson, C.; Barnett, D.; Vaala, W.; Gutierrez, C.; McDaniel, R.; Whitehead, B.; Manning, J. Surveillance programme for important equine infectious respiratory pathogens in the USA. Vet. Rec. 2011, 169, 12. [Google Scholar] [CrossRef] [PubMed]
- Timoney, J.F. The pathogenic equine streptococci. Vet. Res. 2004, 35, 397–409. [Google Scholar] [CrossRef]
- Wilson, W.D. Equine herpesvirus 1 myeloencephalopathy. Vet. Clin. North Am. Equine Pract. 1997, 13, 53–72. [Google Scholar] [CrossRef]
- Van Maanen, C. Equine herpesvirus 1 and 4 infections: An update. Vet. Q. 2002, 24, 57–78. [Google Scholar] [CrossRef]
- Allen, G.; Kydd, J.; Slater, J.; Smith, K. Recent advances in understanding the pathogenesis, epidemiology, and immunological control of equid herpesvirus-1 (EHV-1) abortion. Proceedings of the 8th International Conference on Equine Infectious Diseases. J. Equine Vet. Sci. 1999, 8, 129–146. [Google Scholar]
- Reed, S.M.; Toribio, R.E. Equine herpesvirus 1 and 4. Vet. Clin. North Am. Equine Pract. 2004, 20, 631–642. [Google Scholar] [CrossRef]
- Ostlund, E.N. The equine herpesviruses. Vet. Clin. North Am. Equine Pract. 1993, 9, 283–294. [Google Scholar] [CrossRef]
- Fortier, G.; Van Erck, E.; Pronost, S.; Lekeux, P.; Thiry, E. Equine gammaherpesviruses: Pathogenesis, epidemiology and diagnosis. Vet. J. 2010, 186, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Ficorilli, N.; Studdert, M.; Crabb, B. The nucleotide sequence of Asinine Herpesvirus 3 glycoprotein G indicates that the donkey virus is closely related to equine herpesvirus 1. Arch. Virol. 1995, 140, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.A.; Pusterla, N.; Balasuriya, U.B.; Mapes, S.M.; Nyberg, N.L.; Maclachlan, N.J. Isolation of a gammaherpesvirus similar to asinine herpesvirus-2 (AHV-2) from a mule and a survey of mules and donkeys for AHV-2 infection by real-time PCR. Vet. Microbiol. 2008, 130, 176–183. [Google Scholar] [CrossRef]
- Kleiboeker, S.M.; Schommer, S.K.; Johnson, P.J.; Ehlers, B.; Turnquist, S.E.; Boucher, M.; Kreeger, J.M. Association of two newly recognized herpesviruses with interstitial pneumonia in donkeys (Equus asinus). J. Vet. Diagn. Investig. 2002, 14, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Kleiboeker, S.M.; Turnquist, S.E.; Johnson, P.J.; Kreeger, J.M. Detection and nucleotide sequencing of a DNA-packaging protein gene of equine gammaherpesviruses. J. Vet. Diagn. Investig. 2004, 16, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Thiemann, A. Respiratory disease in the donkey. Equine Vet. Educ. 2012, 24, 469–478. [Google Scholar] [CrossRef]
- Vengust, M.; Wen, X.; Bienzle, D. Herpesvirus-associated neurological disease in a donkey. J. Vet. Diagn. Investig. 2008, 20, 820–823. [Google Scholar] [CrossRef]
- Browning, G.; Ficorilli, N.; Studdert, M. Asinine Herpesvirus genomes: Comparison with those of the equine herpesviruses. Arch. Virol. 1988, 101, 183–190. [Google Scholar] [CrossRef]
- Goodrich, E.L.; Behling-Kelly, E. Clinical Pathology of Donkeys and Mules. Vet. Clin. North Am. Equine Pract. 2019, 35, 433–455. [Google Scholar] [CrossRef]
- Valle, E.; Raspa, F.; Giribaldi, M.; Barbero, R.; Bergagna, S.; Antoniazzi, S.; Mc Lean, A.K.; Minero, M.; Cavallarin, L. A functional approach to the body condition assessment of lactating donkeys as a tool for welfare evaluation. PeerJ 2017, 5, e3001. [Google Scholar] [CrossRef]
- Polidori, P.; Vincenzetti, S. Chapter 4. Farm management and feeding strategies for donkey milk production. In Agricultural Research Updates; Mandhatri, P.G.S., Ed.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2017; p. 93. [Google Scholar]
- Navas González, F.J.; Jordana Vidal, J.; León Jurado, J.M.; Arando Arbulu, A.; McLean, A.K.; Delgado Bermejo, J.V. Genetic parameter and breeding value estimation of donkeys’ problem-focused coping styles. Behav. Processes 2018, 153, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Veterinary Medicine PCR Laboratory, UCDavis. q-PCR diagnostic submission packet. Available online: https://pcrlab.vetmed.ucdavis.edu/sites/g/files/dgvnsk6571/files/inline-files/DiagnosticPacket08.2019.pdf (accessed on 15 March 2020).
- Derrick, B.; Toher, D.; White, P. How to compare the means of two samples that include paired observations and independent observations: A companion to Derrick, Russ, Toher and White (2017). Tutor. Quant. Methods Psychol. 2017, 13, 120–126. [Google Scholar] [CrossRef]
- Moder, K. Alternatives to F-test in one way ANOVA in case of heterogeneity of variances (a simulation study). Psychol. Test Assess. Model. 2010, 52, 343–353. [Google Scholar]
- Rasch, D.; Kubinger, K.D.; Moder, K. The two-sample t test: Pre-testing its assumptions does not pay off. Stat. Pap. 2011, 52, 219–231. [Google Scholar] [CrossRef]
- IBM Corp. IBM SPSS Statistics for Windows; 25.0; IBM Corp: Armonk, NY, USA, 2017. [Google Scholar]
- Derrick, B. The Partially Overlapping Samples t-test. Package ‘Partiallyoverlapping’ for RStudio. Available online: https://cran.r-project.org/web/packages/Partiallyoverlapping/index.html (accessed on 4 May 2020).
- RStudio Team. RStudio 1.1.463; RStudio: Integrated Development for R. RStudio, Inc.: Boston, MA, USA, 2016. [Google Scholar]
- Oliveira, F.G.; Cook, R.F.; Naves, J.H.F.; Oliveira, C.H.S.; Diniz, R.S.; Freitas, F.J.C.; Lima, J.M.; Sakamoto, S.M.; Leite, R.C.; Issel, C.J.; et al. Equine infectious anemia prevalence in feral donkeys from Northeast Brazil. Prev. Vet. Med. 2017, 140, 30–37. [Google Scholar] [CrossRef]
- Hartley, C.A.; Dynon, K.J.; Mekuria, Z.H.; El-Hage, C.M.; Holloway, S.A.; Gilkerson, J.R. Equine gammaherpesviruses: Perfect parasites? Vet. Microbiol. 2013, 167, 86–92. [Google Scholar] [CrossRef]
- Barrandeguy, M.E.; Carossino, M. Infectious diseases in donkeys and mules: An overview and update. J. Equine Vet. Sci. 2018, 65, 98–105. [Google Scholar] [CrossRef]
- Edington, N.; Bridges, C.; Huckle, A. Experimental reactivation of equid herpesvirus 1 (EHV 1) following the administration of corticosteroids. Equine Vet. J. 1985, 17, 369–372. [Google Scholar] [CrossRef]
- Osterrieder, K. Herpesvirales. In Fenner’s Veterinary Virology, 5th ed.; MacLachlan, N.J., Dubovi, E.J., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 189–216. [Google Scholar]
- Rushton, J.O.; Kolodziejek, J.; Nell, B.; Nowotny, N. Prevalence of Asinine Herpesvirus type 5 (AsHV-5) infection in clinically normal Lipizzaner horses. Vet. J. 2014, 200, 200–203. [Google Scholar] [CrossRef]
- Fortier, G.; Van Erck, E.; Fortier, C.; Richard, E.; Pottier, D.; Pronost, S.; Miszczak, F.; Thiry, E.; Lekeux, P. Herpesviruses in respiratory liquids of horses: Putative implication in airway inflammation and association with cytological features. Vet. Microbiol. 2009, 139, 34–41. [Google Scholar] [CrossRef][Green Version]
- Goehring, L.S. Donkeys. In Robinson’s Current Therapy in Equine Medicine, 1st ed.; Sprayberry, K.A., Robinson, N.E., Eds.; Elsevier: Philadelphia, PA, USA, 2015; pp. 155–157. [Google Scholar]
- Rickards, K.J.; Thiemann, A.K. Respiratory Disorders of the Donkey. Vet. Clin. N. Am. Equine Pract. 2019, 35, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Laus, F.; Preziuso, S.; Spaterna, A.; Beribe, F.; Tesei, B.; Cuteri, V. Clinical and epidemiological investigation of chronic upper respiratory diseases caused by beta-haemolytic Streptococci in horses. Comp. Immunol. Microbiol. Infect. Dis. 2007, 30, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Gao, N.; Waller, A.; Cook, F.; Fan, S.; Yuan, D.; Du, Y.; Li, F.; Norimine, J.; Zhu, W. An outbreak of strangles associated with a novel genotype of Streptococcus equi subspecies equi in donkeys in China during 2018. Equine Vet. J. 2019, 51, 743–748. [Google Scholar] [CrossRef] [PubMed]

| Assay Name | Gene, NCBI a Accession # | Assay Location (bp) | Amplicon Length (bp) |
|---|---|---|---|
| Asinine Herpesvirus 2 | Polymerase, EU165547 | 100 | 81 |
| Asinine Herpesvirus 3 | Glycoprotein B, U24184 | 740 | 145 |
| Asinine Herpesvirus 5 | Polymerase, AY054993 | 600 | 64 |
| Equine Herpesvirus 1 | Glycoprotein B, NC_001491 | 400 | 89 |
| Equine Herpesvirus 1, neuropathogenic | ORF 30, KF644574 | 200 | 92 |
| Equine Herpesvirus 1, non-neuropathogenic | ORF 30, KX101095 | 200 | 92 |
| Equine Herpesvirus 4 | Glycoprotein B, AF030027 | 440 | 77 |
| Streptococcus equi subspecies equi | M Protein, AF012927 | 150 | 185 |
| Streptococcus equi subspecies zooepidemicus | ITS, EU860336 | 80 | 88 |
| Influenza A (H3N8) | Hemagglutinin Precursor, EF541443 | 350 | 200 |
| Equine rhinitis A virus | RNA polymerase, X96870 | 150 | 111 |
| Equine rhinitis B virus | RNA polymerase, X96871 | 350 | 87 |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH, AF097179 | 60 | 105 |
| Parameters | Mean | SEM | SD | Skewness | Kurtosis | |
|---|---|---|---|---|---|---|
| Clinical Examination Signs | Body condition score (BCS) | 3.12 | 0.09 | 0.83 | −0.15 | −0.96 |
| Behavior signs | 2.11 | 0.04 | 0.38 | 1.14 | 3.13 | |
| Skin/hair condition | 1.29 | 0.10 | 0.94 | 3.21 | 9.14 | |
| Lameness presence | 1.96 | 0.02 | 0.19 | −5.13 | 24.88 | |
| Ocular discharge presence | 1.82 | 0.04 | 0.38 | −1.73 | 1.01 | |
| Nasal discharge presence | 1.66 | 0.05 | 0.48 | −0.68 | −1.57 | |
| Abnormal breathing presence | 1.07 | 0.03 | 0.26 | 3.41 | 9.88 | |
| Coughing presence | 1.93 | 0.03 | 0.26 | −3.41 | 9.88 | |
| SAA | SAA maximal concentration (mg/L) | 10.80 | 3.72 | 34.27 | 5.42 | 33.75 |
| Pathogen Load qPCR-Assay | Asinine Herpesvirus 2 (AHV-2) aka EHV7 | 39.69 | 0.14 | 1.30 | −4.33 | 18.30 |
| Asinine Herpesvirus 3 (AHV-3) aka EHV8 | 39.74 | 0.14 | 1.30 | −5.92 | 36.83 | |
| Asinine Herpesvirus 5 (AHV-5) | 34.39 | 0.50 | 4.59 | −0.51 | −0.38 | |
| Equine Herpesvirus 1 (EHV-1) | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine Herpesvirus 1 (EHV-1) neuropathogenic | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine Herpesvirus 1 (EHV-1) non-neuropathogenic | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine herpesvirus 4 (EHV-4) | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Streptococcus equi subspecies equi | 39.94 | 0.04 | 0.39 | −6.77 | 46.45 | |
| Streptococcus equi subspecies zooepidemicus | 35.04 | 0.44 | 4.08 | -0.75 | 0.07 | |
| Influenza AH3N8 | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine rhinitis A virus | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine rhinitis B virus | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Controls | Glyceraldehyde 3 phosphate dehydrogenase (First control) | 29.24 | 0.38 | 3.49 | 0.83 | 1.91 |
| Glyceraldehyde 3 phosphate dehydrogenase (Second control) | 32.13 | 0.41 | 3.78 | 0.27 | −0.87 | |
| Fibrinogen | 0.44 | 0.09 | 0.50 | 4.56 | 23.39 |
| Parameters | Mean | SEM | SD | Skewness | Kurtosis | |
|---|---|---|---|---|---|---|
| Clinical Examination Signs | Body condition score | 2.43 | 0.08 | 0.37 | 0.97 | 1.81 |
| Behavior signs | 1.96 | 0.07 | 0.36 | −0.65 | 6.34 | |
| Skin/Hair condition | 1.00 | 0.00 | 0.00 | N/A | N/A | |
| Lameness presence | 2.00 | 0.00 | 0.00 | N/A | N/A | |
| Ocular discharge presence | 1.87 | 0.07 | 0.34 | −2.42 | 4.21 | |
| Nasal discharge presence | 1.42 | 0.10 | 0.50 | 0.36 | −2.05 | |
| Abnormal breathing presence | 1.13 | 0.07 | 0.34 | 2.42 | 4.21 | |
| Coughing presence | 1.13 | 0.07 | 0.34 | 2.42 | 4.21 | |
| SAA | SAA maximal concentration (mg/L) | 17.42 | 10.73 | 52.58 | 4.43 | 20.53 |
| Pathogen Load qPCR-Assay | Asinine Herpesvirus 2 (AHV-2) aka EHV7 | 38.03 | 0.90 | 4.40 | −2.57 | 6.18 |
| Asinine Herpesvirus 3 (AHV-3) aka EHV8 | 39.94 | 0.06 | 0.31 | −4.90 | 24.00 | |
| Asinine Herpesvirus 5 (AHV-5) | 32.65 | 0.89 | 4.36 | −1.37 | 2.84 | |
| Equine Herpesvirus 1 (EHV-1) | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine Herpesvirus 1 (EHV-1) neuropathogenic | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine Herpesvirus 1 (EHV-1) non-neuropathogenic | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine Herpesvirus 4 (EHV-4) | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Streptococcus equi subspecies equi | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Streptococcus equi subspecies zooepidemicus | 33.04 | 0.69 | 3.38 | -0.65 | 0.77 | |
| Influenza AH3N8 | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine rhinitis A virus | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Equine rhinitis B virus | 40.00 | 0.00 | 0.00 | N/A | N/A | |
| Controls | Glyceraldehyde 3 phosphate dehydrogenase (First control) | 23.61 | 0.97 | 4.76 | 2.04 | 5.02 |
| Glyceraldehyde 3 phosphate dehydrogenase (Second control) | 29.37 | 0.85 | 4.17 | 1.45 | 1.75 | |
| Fibrinogen | 0.63 | 0.54 | 2.67 | 4.73 | 22.74 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jerele, S.; Davis, E.; Mapes, S.; Pusterla, N.; Navas González, F.J.; Iglesias Pastrana, C.; Abdelfattah, E.M.; McLean, A. Survey of Serum Amyloid A and Bacterial and Viral Frequency Using qPCR Levels in Recently Captured Feral Donkeys from Death Valley National Park (California). Animals 2020, 10, 1086. https://doi.org/10.3390/ani10061086
Jerele S, Davis E, Mapes S, Pusterla N, Navas González FJ, Iglesias Pastrana C, Abdelfattah EM, McLean A. Survey of Serum Amyloid A and Bacterial and Viral Frequency Using qPCR Levels in Recently Captured Feral Donkeys from Death Valley National Park (California). Animals. 2020; 10(6):1086. https://doi.org/10.3390/ani10061086
Chicago/Turabian StyleJerele, Sara, Eric Davis, Samantha Mapes, Nicola Pusterla, Francisco Javier Navas González, Carlos Iglesias Pastrana, Essam Mahmoud Abdelfattah, and Amy McLean. 2020. "Survey of Serum Amyloid A and Bacterial and Viral Frequency Using qPCR Levels in Recently Captured Feral Donkeys from Death Valley National Park (California)" Animals 10, no. 6: 1086. https://doi.org/10.3390/ani10061086
APA StyleJerele, S., Davis, E., Mapes, S., Pusterla, N., Navas González, F. J., Iglesias Pastrana, C., Abdelfattah, E. M., & McLean, A. (2020). Survey of Serum Amyloid A and Bacterial and Viral Frequency Using qPCR Levels in Recently Captured Feral Donkeys from Death Valley National Park (California). Animals, 10(6), 1086. https://doi.org/10.3390/ani10061086