Analysis of the Bovine DLK1 Gene Polymorphism and Its Relation to Lipid Metabolism in Chinese Simmental
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.1.1. Cell Line
2.1.2. Animals and Traits Analysis
2.2. Construction of PGPU6-shDLK1 and PBI-CMV3-DLK1 Vectors
2.3. BFFs Culture and Transfection
2.4. Analysis of mRNA Levels of DLK1 and Lipid Metabolism-Related Genes
2.5. Analysis of Protein Levels of DLK1
2.6. Determination of TGs Content in BFFs
2.7. Determination of Fatty Acid Contents in BFFs
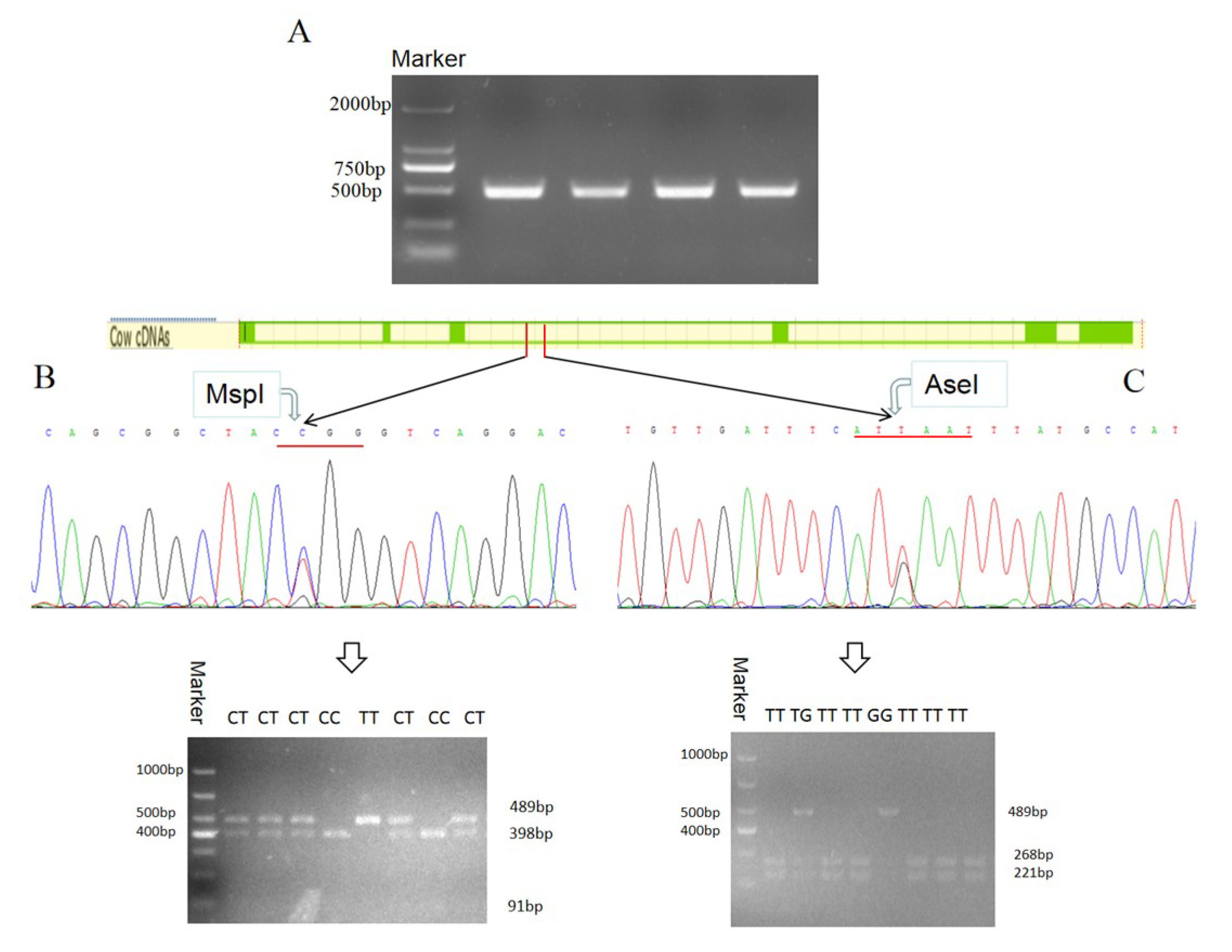
2.8. The Polymorphism of Bovine DLK1 Gene
2.9. Statistical Analysis
3. Results
3.1. Bovine DLK1 Gene Regions’ Location and Transcripts
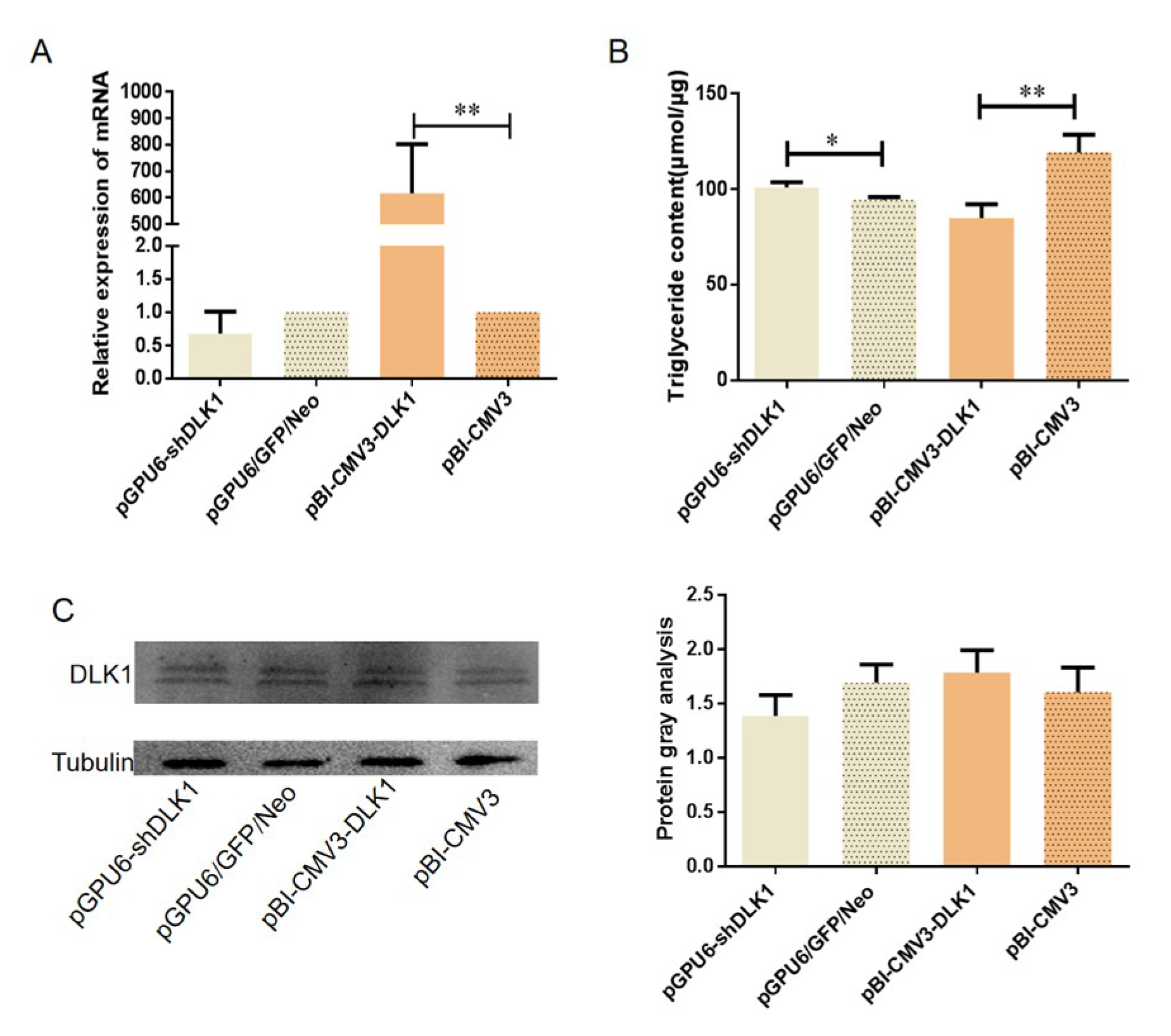
3.2. The Expression of the DLK1 Gene in BFFs after Overexpression or Interference
3.3. The mRNA Expression of Genes Related to Lipid Metabolism in BFFs after Interference and Overexpression of Bovine DLK1
3.4. Determination of the Fatty Acid Content in BFFs after Interference and Overexpression of the DLK1 Gene
3.5. Polymorphism of Bovine DLK1 Gene and Its Association with Economic Traits in Chinese Simmental Steers
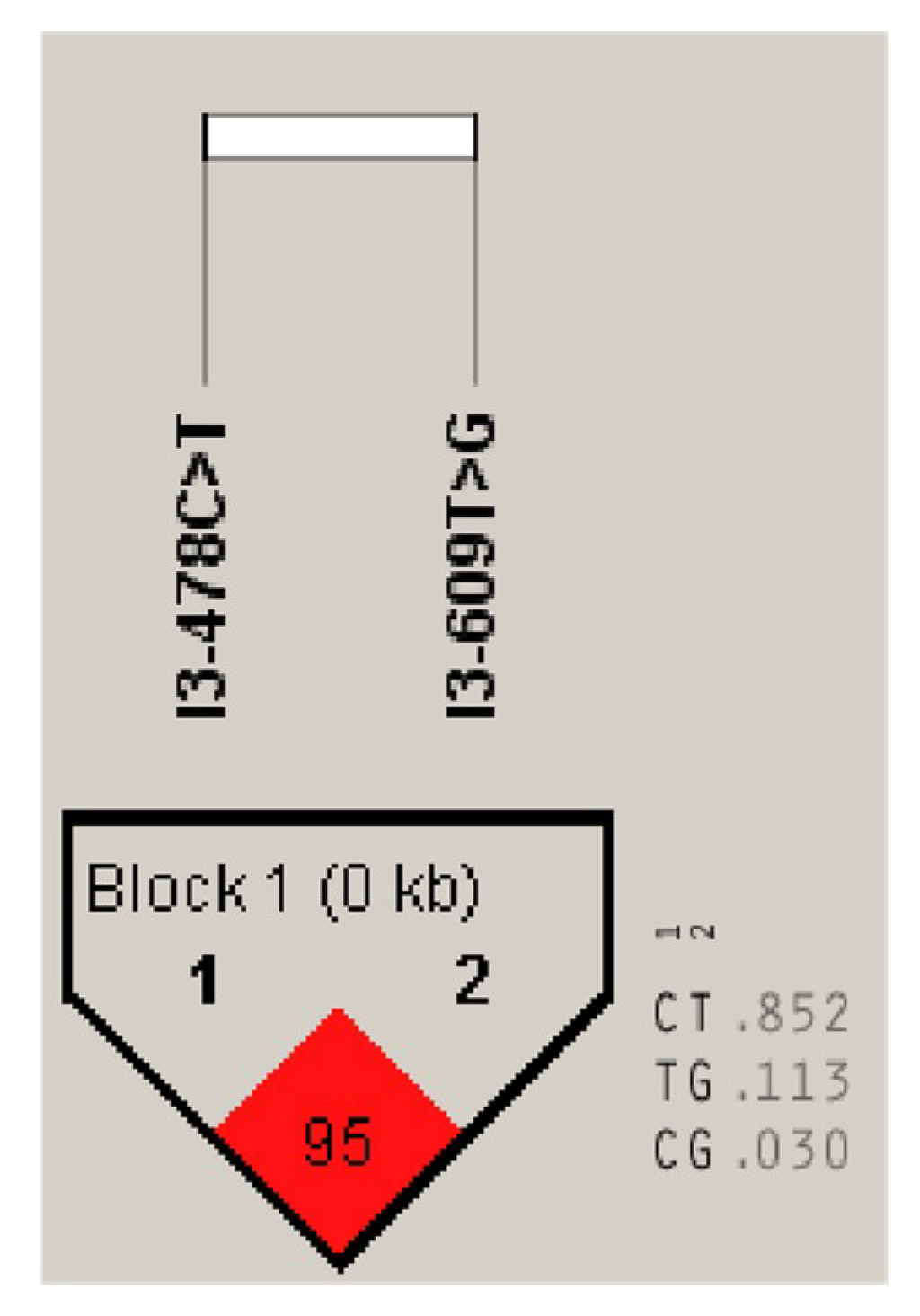
3.5.1. Two SNPs of the DLK1 Gene of Chinese Simmental Steers
3.5.2. Genetic Diversity of the DLK1 Gene in Chinese Simmental Steers Population
3.5.3. Association of DLK1 Gene Polymorphisms with the Carcass and Meat Quality Traits in Chinese Simmental
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Garcés, C.; Ruiz-Hidalgo, M.J.; Bonvini, E.; Goldstein, J.; Laborda, J. Adipocyte differentiation is modulated by secreted delta-like (dlk) variants and requires the expression of membrane-associated dlk. Differentiation 2010, 64, 103–114. [Google Scholar] [CrossRef]
- Smas, C.M.; Sul, H.S. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 1993, 73, 725–734. [Google Scholar] [CrossRef]
- Moon, Y.S.; Smas, C.M.; Lee, K.; Villena, J.A.; Kim, K.H.; Yun, E.J.; Sul, H.S. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol. Cell. Biol. 2002, 22, 5585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, E.; Jensen, C.H.; Schrøder, H.D.; Farnir, F.; Shay-Hadfield, T.; Kliem, A.; Cockett, N.; Georges, M.; Charlier, C. Ectopic expression of DLK1 protein in skeletal muscle of padumnal heterozygotes causes the callipyge phenotype. Iowa State University. Curr. Biol. 2004, 14, 1858. [Google Scholar] [CrossRef] [Green Version]
- Shin, J. The Role of Chicken Delta-Like Protein 1 Expression in Skeletal Muscle Development and Regeneration. Bachelor’s Thesis, Iowa State University, AAmes, IA, USA, 2009. [Google Scholar]
- Albrecht, E.; Kuzinski, J.; Komolka, K.; Gotoh, T.; Maak, S. Localization and abundance of early markers of fat cell differentiation in the skeletal muscle of cattle during growth—Are DLK1-positive cells the origin of marbling flecks? Meat Sci. 2015, 100, 237–245. [Google Scholar] [CrossRef]
- Kim, K.S.; Kim, J.-J.; Dekkers, J.C.M.; Rothschild, M.F. Polar overdominant inheritance of a DLK1 polymorphism is associated with growth and fatness in pigs. Mamm. Genome 2004, 15, 552–559. [Google Scholar] [CrossRef]
- Chen, F.Y.; Niu, H.; Lei, C.Z.; Lan, X.Y.; Zhang, C.L.; Li, M.J.; Hua, L.S.; Chen, H.; Wang, J.Q.; Wang, J. Polymorphism of DLK1 and CLPG gene and their association with phenotypic traits in Chinese cattle. Mol. Boil. Rep. 2010, 38, 243–248. [Google Scholar]
- Magee, D.A.; Berry, D.P.; Berkowicz, E.W.; Sikora, K.M.; Howard, D.J.; Mullen, M.P.; Evans, R.D.; Spillane, C.; MacHugh, D.E. Single Nucleotide Polymorphisms within the Bovine DLK1-DIO3 Imprinted Domain Are Associated with Economically Important Production Traits in Cattle. J. Hered. 2010, 102, 94–101. [Google Scholar]
- Yu, H.; Zhao, Z.; Yu, X.; Li, J.; Lu, C.; Yang, R. Bovine lipid metabolism related gene GPAM: Molecular characterization, function identification, and association analysis with fat deposition traits. Gene 2017, 609, 9–18. [Google Scholar]
- Fang, X.B.; Zhang, L.P.; Yu, X.Z.; Li, J.Y.; Lu, C.Y.; Zhao, Z.; Yang, R. Association of HSL gene E1-c.276C > T and E8-c.51C > T mutation with economical traits of Chinese Simmental cattle. Mol. Boil. Rep. 2014, 41, 105. [Google Scholar] [CrossRef]
- Fahrenkrug, S.C.; Freking, B.; Smith, T.P. Genomic Organization and Genetic Mapping of the Bovine PREF-1 Gene. Biochem. Biophys. Res. Commun. 1999, 264, 662–667. [Google Scholar] [PubMed] [Green Version]
- Vuocolo, T.; Pearson, R.; Campbell, P.; Tellam, R.L. Differential expression of Dlk-1 in bovine adipose tissue depots. Comp. Biochem. Physiol. Part B Biochem. Mol. Boil. 2003, 134, 315–333. [Google Scholar] [CrossRef]
- Greenbaum, D.; Colangelo, C.; Williams, K.; Gerstein, M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Boil. 2003, 4, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christy, R.J.; Kaestner, K.H.; Geiman, D.E.; Lane, M.D. CCAAT/enhancer binding protein gene promoter: Binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. USA 1991, 88, 2593–2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kersten, S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001, 2, 282–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, I.J. Lipoprotein lipase and lipolysis: Central roles in lipoprotein metabolism and atherogenesis. J. Lipid Res. 1996, 37, 693–707. [Google Scholar] [PubMed]
- Stein, D.T.; Stevenson, E.B.; Chester, M.W.; Basit, M.; Daniels, M.B.; Turley, S.D.; McGarry, J.D. The insulinotropic potency of fatty acids is influenced profoundly by their chain length and degree of saturation. J. Clin. Investig. 1997, 100, 398–403. [Google Scholar]
- Sergeant, S.; Rahbar, E.; Chilton, F.H. Gamma-linolenic acid, Dihommo-gamma linolenic, Eicosanoids and Inflammatory Processes. Eur. J. Pharmacol. 2016, 785, 77–86. [Google Scholar]
- Phinney, S.D. Arachidonic acid maldistribution in obesity. Lipids 1996, 31, S271–S274. [Google Scholar]
- Spector, A.A.; Kim, H.-Y. Discovery of essential fatty acids. J. Lipid Res. 2014, 56, 11–21. [Google Scholar]
- Charalambous, M.; Da Rocha, S.T.; Radford, E.J.; Medina-Gomez, G.; Curran, S.; Pinnock, S.B.; Ferguson-Smith, A.C. DLK1/PREF1 regulates nutrient metabolism and protects from steatosis. Proc. Natl. Acad. Sci. 2014, 111, 16088–16093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flehmig, G.; Scholz, M.; Klöting, N.; Fasshauer, M.; Tonjes, A.; Stumvoll, M.; Youn, B.-S.; Blüher, M. Identification of Adipokine Clusters Related to Parameters of Fat Mass, Insulin Sensitivity and Inflammation. PLoS ONE 2014, 9, e99785. [Google Scholar]

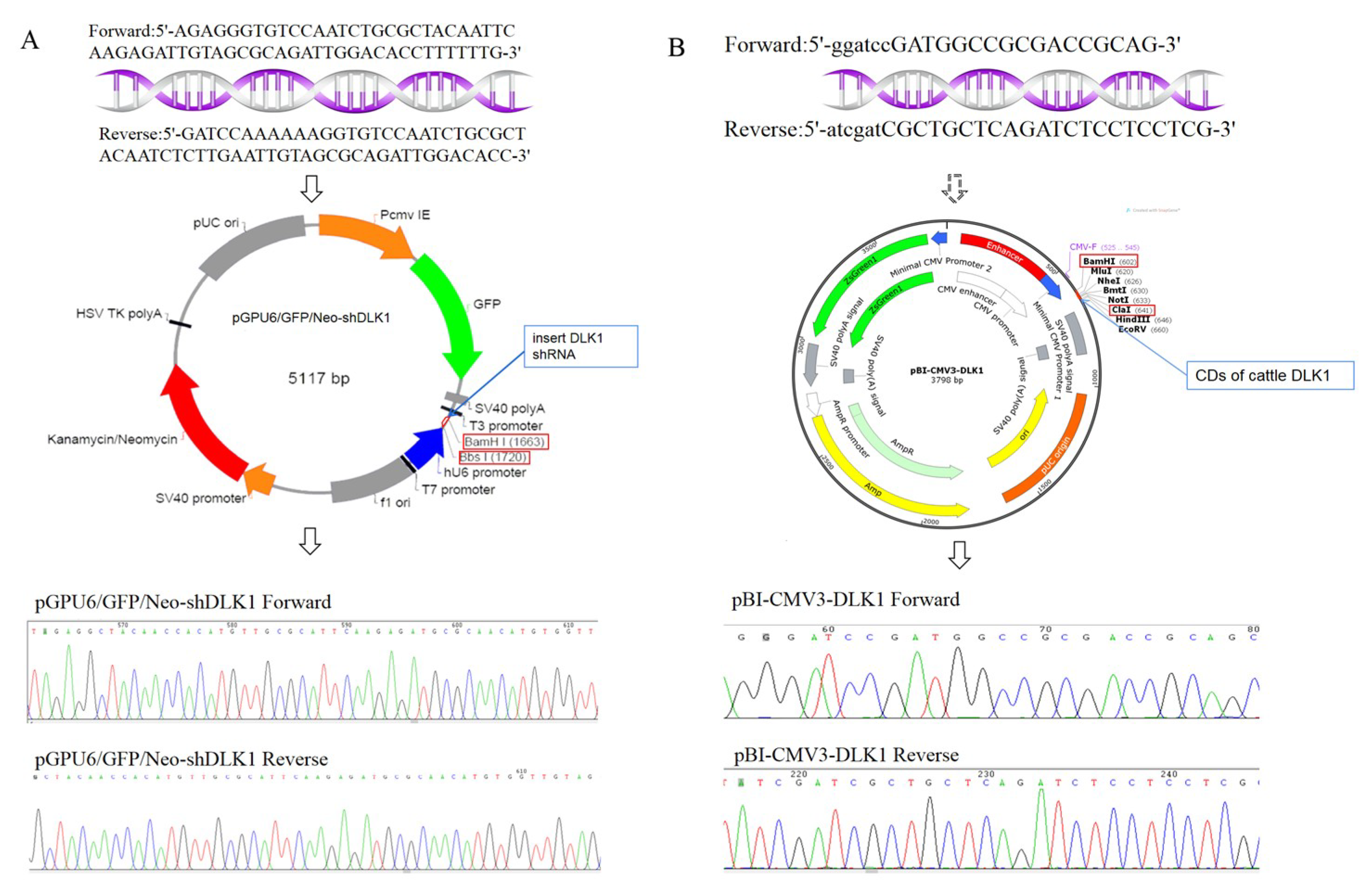



| Primer | Forward Sequences (5′-3′) | Reverse Sequences (5′-3′) | Target Sequences |
|---|---|---|---|
| shRNA of bovine DLK1 | AGAGGCTACAACCACATGTTGCGCATTCAAGAGATGCGCAACATGTGGTTGTAGCTTTTTTG | GATCCAAAAAAGCTACAACCACATGTTGCGCATCTCTTGAATGCGCAACATGTGGTTGTAGC | GCTACAACCACATGTTGCGCA |
| surpass coding sequence of bovine DLK1 | TCCGCAACCAGAAGCCCA | GAGCGTAGCGTTCACCAGATTT | |
| coding sequence of bovine DLK1 | GGATCCGATGGCCGCGACCGCAG | ATCGATCGCTGCTCAGATCTCCTCCTCG | |
| quantitative primer of bovine DLK1 | CTTGCTCCTGCTGGCTTTCG | AGGTCACGCACTGGTCACAC | |
| quantitative primer of bovine LPL | CCGCAGACAGGATTACAG | GTGGTTGAAGTGACAGTTAG | |
| quantitative primer of bovine PPARγ | CCTTCACCACCGTTGACTTCTC | GATACAGGCTCCACTTTGATTGC | |
| quantitative primer of bovine C/EBPα | CCGTGGACAAGAACAAGCAAC | TGGTCAGCTCCAGCACCTTC | |
| Polymorphism primer of bovine DLK1 | TCCACAGGTGAGGCTACTAAG | CTGTTCTCCTGACTTCCTAAG | |
| β-actin | AGAGCAAGAGAGGCATCC | TCGTTGTAGAAGGTGTGGT |
| SNP | IVS3 + 478 C > T(n = 237) | IVS3 + 609 T > G(n = 217) | ||
|---|---|---|---|---|
| Location | Intron3 | Intron3 | ||
| Gene frequency | C T | 0.865 0.135 | T G | 0.846 0.154 |
| Genotype frequency | CC CT TT | 0.738 0.253 0.008 | TT TG GG | 0.719 0.234 0.047 |
| PIC a | 0.21 | 0.23 | ||
| Ho b | 0.75 | 0.77 | ||
| He c | 0.25 | 0.23 | ||
| Economical Traits | Genotypes of IVS3 + 478 C > T | Genotypes of IVS3 + 609 T > G | |||
|---|---|---|---|---|---|
| CC | CT | TT | TG | GG | |
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |
| LW (kg) | 486.89 ± 56.00 | 481.99 ± 58.85 | 489.13 ± 58.37 | 486.72 ± 62.82 | 488.67 ± 68.85 |
| CW (kg) | 253.47 ± 35.00 | 249.99 ± 36.90 | 255.16 ± 35.89 | 252.48 ± 3 9.97 | 252.58 ± 46.58 |
| KFW (kg) | 4.44 ± 2.72 a | 5.07 ± 2.89 b | 4.53 ± 2.77 a | 4.96 ± 2.88 a | 3.46 ± 2.70 b |
| CL (cm) | 140.45 ± 9.01 a | 138.14 ± 6.53 b | 140.19 ± 8.96 a | 138.79±6.94 b | 145.83±4.96 |
| TL (cm) | 6.70 ± 0.87 | 6.93 ± 0.89 | 6.72 ± 0.88 | 6.92 ± 0.90 | 6.55 ± 1.03 |
| CD (cm) | 64.42 ± 3.37 | 64.45 ± 3.19 | 64.46 ± 3.32 a | 64.57 ± 3.28 a | 67.50 ± 1.87 b |
| BFT (cm) | 0.92 ± 0.64 a | 1.05 ± 0.65 b | 0.94 ± 0.65 a | 1.05 ± 0.63 a | 0.70 ± 0.61 b |
| FCR (%) | 46.64 ± 22.16 a | 51.37 ± 20.46 b | 47.50 ± 22.04 AB | 52.29 ± 21.13 A | 39.17 ± 23.54 B |
| MBS | 5.45 ± 0.67 a | 5.20 ± 0.80 b | 5.41 ± 0.67 | 5.36 ± 0.75 | 5.00 ± 1.10 |
| LEA (cm2) | 78.50 ± 12.99 a | 75.17 ± 10.32 b | 79.09 ± 13.24 | 74.62 ± 10.22 | 78.50 ± 12.15 |
| FCS | 2.87 ± 0.94 A | 2.50 ± 0.98 B | 2.83 ± 0.94 AB | 2.42 ± 0.96 B | 3.33 ± 0.52 A |
| Types of Fatty Acids | Genotypes (IVS3 + 478 C > T) | Genotypes (IVS3 + 609 T > G) | |||
|---|---|---|---|---|---|
| CC | CT | TT | TG | GG | |
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |
| Myristic acid (c14:0) | 0.020 ± 0.019 | 0.028 ± 0.021 | 0.020 ± 0.017 | 0.027 ± 0.023 | 0.024 ± 0.009 |
| Myristic oleic acid (c14:1) | 0.002 ± 0.006 | 0.004 ± 0.004 | 0.002 ± 0.004 | 0.004 ± 0.004 | 0.001 ± 0.002 |
| Hexadecanoic acid (c16:0) | 0.263 ± 0.222 | 0.341 ± 0.200 | 0.251 ± 0.175 | 0.332 ± 0.223 | 0.277 ± 0.087 |
| Palmitoleic acid (c16:1) | 0.029 ± 0.038 | 0.035 ± 0.023 | 0.026 ± 0.020 | 0.033 ± 0.026 | 0.023 ± 0.013 |
| Margaric acid (c17:0) | 0.012 ± 0.008 | 0.015 ± 0.007 | 0.012 ± 0.007 | 0.014 ± 0.008 | 0.011 ± 0.003 |
| Heptadecenoic acid (c17:1) | 0.005 ± 0.008 | 0.007 ± 0.006 | 0.005 ± 0.006 | 0.006 ± 0.006 | 0.005 ± 0.006 |
| Stearic acid (c18:0) | 0.189±0.120 | 0.241 ± 0.118 | 0.184 ± 0.110 | 0.235 ± 0.133 | 0.219 ± 0.044 |
| Oleic acid (c18:1n9c) | 0.389 ± 0.469 | 0.424 ± 0.230 | 0.346 ± 0.236 | 0.416 ± 0.255 | 0.349 ± 0.113 |
| Linoleic acid (c18:2n6c) | 0.098 ± 0.028 b | 0.118 ± 0.050 a | 0.096 ± 0.026 b | 0.120 ± 0.053 a | 0.098 ± 0.023 |
| α-Linolenic acid (c18:3n3) | 0.005 ± 0.006 | 0.009 ± 0.012 | 0.004 ± 0.005 | 0.009 ± 0.013 | 0.009 ± 0.002 |
| Arachidic acid (c20:0) | 0.000 ± 0.001 | 0.001 ± 0.002 | 0.000 ± 0.001 | 0.001 ± 0.002 | 0.000 ± 0.001 |
| Eicosenoic acid (c20:1) | 0.001 ± 0.003 | 0.001 ± 0.001 | 0.000 ± 0.001 | 0.000 ± 0.001 | 0.000 ± 0.001 |
| Dohono-γ-linolenic acid (c20:3n6) | 0.010 ± 0.002 | 0.010 ± 0.004 | 0.010 ± 0.002 | 0.009 ± 0.004 | 0.009 ± 0.002 |
| Arachidonic acid (c20:4n6) | 0.048 ± 0.011 | 0.056 ± 0.019 | 0.048 ± 0.010 b | 0.055 ± 0.020 a | 0.043 ± 0.005 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Jiang, P.; Yu, X.; Mi, J.; Bai, Z.; Zhang, X.; Liu, Y.; Fang, X.; Yang, R.; Zhao, Z. Analysis of the Bovine DLK1 Gene Polymorphism and Its Relation to Lipid Metabolism in Chinese Simmental. Animals 2020, 10, 923. https://doi.org/10.3390/ani10060923
Wang M, Jiang P, Yu X, Mi J, Bai Z, Zhang X, Liu Y, Fang X, Yang R, Zhao Z. Analysis of the Bovine DLK1 Gene Polymorphism and Its Relation to Lipid Metabolism in Chinese Simmental. Animals. 2020; 10(6):923. https://doi.org/10.3390/ani10060923
Chicago/Turabian StyleWang, Mengyan, Ping Jiang, Xiang Yu, Jiaqi Mi, Zitong Bai, Xiuqi Zhang, Yinuo Liu, Xibi Fang, Runjun Yang, and Zhihui Zhao. 2020. "Analysis of the Bovine DLK1 Gene Polymorphism and Its Relation to Lipid Metabolism in Chinese Simmental" Animals 10, no. 6: 923. https://doi.org/10.3390/ani10060923
APA StyleWang, M., Jiang, P., Yu, X., Mi, J., Bai, Z., Zhang, X., Liu, Y., Fang, X., Yang, R., & Zhao, Z. (2020). Analysis of the Bovine DLK1 Gene Polymorphism and Its Relation to Lipid Metabolism in Chinese Simmental. Animals, 10(6), 923. https://doi.org/10.3390/ani10060923





