Diet with a High Proportion of Rice Alters Profiles and Potential Function of Digesta-Associated Microbiota in the Ileum of Goats
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Diets and Management
2.2. Measurements and Analytical Methods
2.3. Bacterial Data Processing and Function Predication
2.4. Metabolites Measured in the Ileal Digesta and Ileal Morphology
2.5. Statistical Analyses
3. Results
3.1. Ileal Bacterial Diversity and Similarity
3.2. Intestinal Bacterial Community Structure
3.3. Function Prediction of Ileal Microbiota Using Picrust
3.4. Metabolites and Biochemical Parameters in the Ileal Digesta
3.5. Relationship among the Bacterial Community and Metabolites and Biochemical Indices
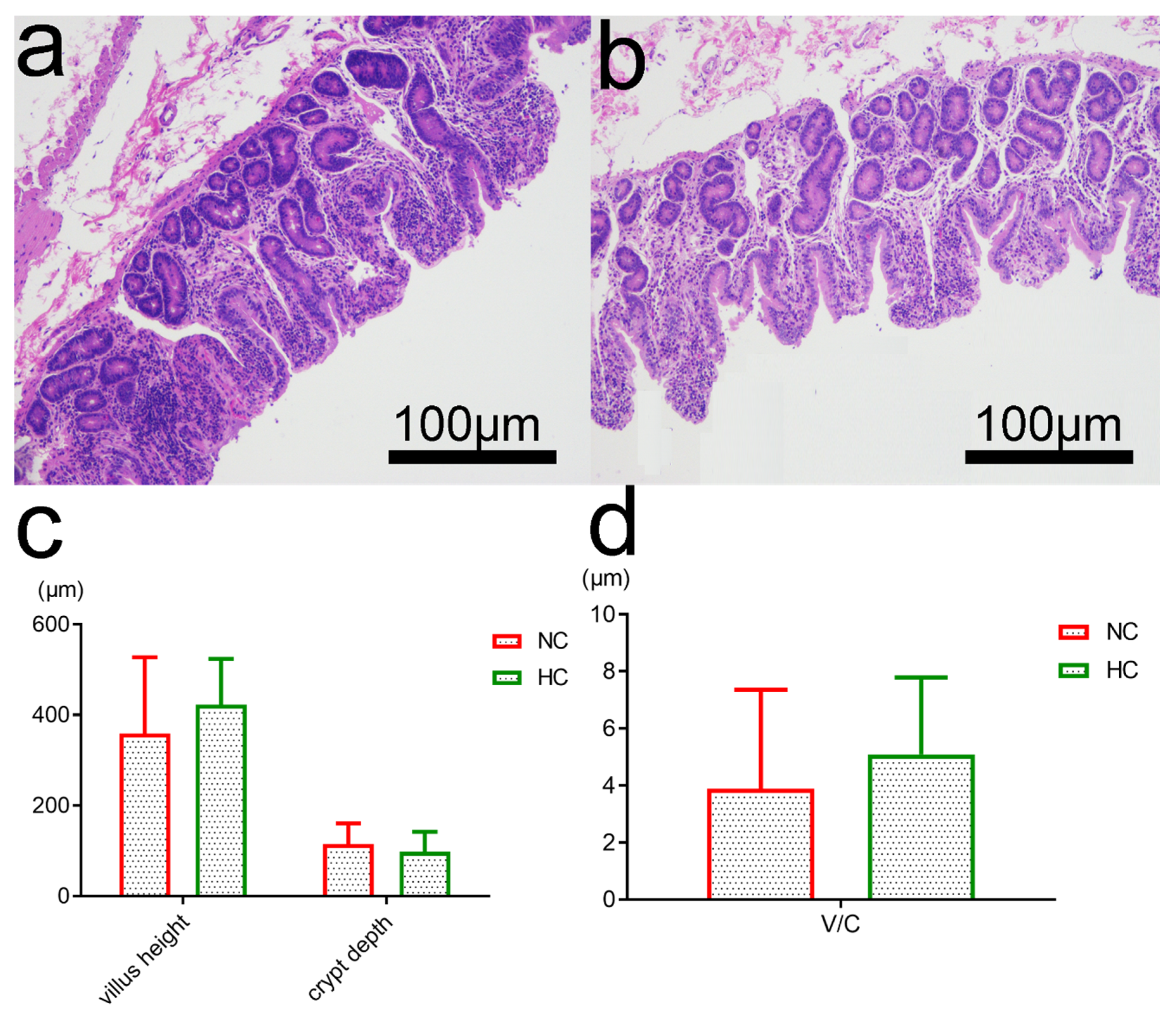
3.6. Intestinal Morphology
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pourazad, P.; Khiaosa-Ard, R.; Qumar, M.; Wetzels, S.U.; Klevenhusen, F.; Metzler-Zebeli, B.U.; Zebeli, Q. Transient feeding of a concentrate-rich diet increases the severity of subacute ruminal acidosis in dairy cattle. J. Anim. Sci. 2016, 94, 726–738. [Google Scholar] [CrossRef]
- Mao, S.Y.; Huo, W.J.; Zhu, W.Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 2016, 18, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.Y.; Ye, H.M.; Liu, J.H.; Mao, S.Y. High-grain diets altered rumen fermentation and epithelial bacterial community and resulted in rumen epithelial injuries of goats. Appl. Microbiol. Biotechnol. 2017, 101, 6981–6992. [Google Scholar] [CrossRef] [PubMed]
- Boerman, J.P.; Potts, S.; VandeHaar, M.J.; Allen, M.S.; Lock, A.L. Milk production responses to a change in dietary starch concentration vary by production level in dairy cattle. J. Dairy Sci. 2015, 98, 4698–4706. [Google Scholar] [CrossRef] [PubMed]
- Zebeli, Q.; Ametaj, B.N. Relationships between rumen lipopolysaccharide and mediators of inflammatory response with milk fat production and efficiency in dairy cows. J. Dairy Sci. 2009, 92, 3800–3809. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.C.; Khafipour, E.; Li, S.; Gozho, G.N.; Krause, D.O. Subacute ruminal acidosis (SARA), endotoxins and health consequences. Anim. Feed Sci. Technol. 2012, 172, 9–21. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Krause, D.O.; Gozho, G.N.; McBride, B.W. Subacute ruminal acidosis in dairy cows: The physiological causes, incidence and consequences. Vet. J. 2008, 176, 21–31. [Google Scholar] [CrossRef]
- Mao, S.Y.; Zhang, R.Y.; Wang, D.S.; Zhu, W.Y. Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe 2013, 24, 12–19. [Google Scholar] [CrossRef]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Changes in the Rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl. Environ. Microbiol. 2013, 79, 3744–3755. [Google Scholar] [CrossRef]
- Collins, S.M.; Surette, M.; Bercik, P. The interplay between the intestinal microbiota and the brain. Nat. Rev. Microbiol. 2012, 10, 735–742. [Google Scholar] [CrossRef]
- Hooper, L.V.; Macpherson, A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010, 10, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Hase, K. Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 2014, 10, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Gressley, T.F.; Hall, M.B.; Armentano, L.E. Ruminant nutrition symposium: Productivity, digestion, and health responses to hindgut acidosis in ruminants. J. Anim. Sci. 2011, 89, 1120–1130. [Google Scholar] [CrossRef]
- Ye, H.M.; Liu, J.H.; Feng, P.F.; Zhu, W.Y.; Mao, S.Y. Grain-rich diets altered the colonic fermentation and mucosa-associated bacterial communities and induced mucosal injuries in goats. Sci. Rep. 2016, 6, 20329. [Google Scholar] [CrossRef]
- Wang, K.J.; Zheng, M.L.; Ren, A.; Zhou, C.S.; Yan, Q.X.; Tan, Z.L.; Zhang, P.H.; Yi, K.L. Effects of high rice diet on growth performance, nutrients apparent digestibility, nitrogen metabolism, blood parameters and rumen fermentation in growing goats. Kafkas Univ. Vet. Fak. Derg. 2019, 25, 749–755. [Google Scholar]
- Jiao, J.Z.; Zhang, X.L.; Wang, M.; Zhou, C.S.; Yan, Q.X.; Tan, Z.L. Linkages between epithelial microbiota and host transcriptome in the ileum during high-grain challenges: Implications for gut homeostasis in goats. J. Agric. Food Chem. 2019, 67, 551–561. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, X.G.; Liao, H.Y.; Tan, Z.L.; Tang, S.X.; Sun, Z.H.; Zhou, C.S.; Han, X.F. Effects of rice straw particle size on digesta particle size distribution, nitrogen metabolism, blood biochemical parameters, microbial amino acid composition and intestinal amino acid digestibility in goats. Anim. Sci. J. 2011, 82, 78–85. [Google Scholar] [CrossRef]
- Kraler, M.; Ghanbari, M.; Domig, K.J.; Schedle, K.; Kneifel, W. The intestinal microbiota of piglets fed with wheat bran variants as characterised by 16S rRNA next generation amplicon sequencing. Arch. Anim. Nutr. 2016, 70, 173–189. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, L.; Liu, J.H.; Zhu, W.Y.; Mao, S.Y. A high grain diet dynamically shifted the composition of mucosa-associated microbiota and induced mucosal injuries in the colon of sheep. Front. Microbiol. 2017, 8, 2080. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.Z.; Huang, J.Y.; Zhou, C.S.; Tan, Z.L. Taxonomic identification of ruminal epithelial bacterial diversity during rumen development in goats. Appl. Environ. Microbiol. 2015, 81, 3502–3509. [Google Scholar] [CrossRef]
- Hua, C.F.; Tian, J.; Tian, P.; Cong, R.H.; Luo, Y.W.; Geng, Y.L.; Zhao, R.Q. Feeding a high concentration diet induces unhealthy alterations in the composition and metabolism of ruminal microbiota and host response in a goat model. Front. Microbiol. 2017, 8, 138. [Google Scholar] [CrossRef]
- Liu, J.H.; Xue, C.X.; Sun, D.M.; Zhu, W.Y.; Mao, S.Y. Impact of high-grain diet feeding on mucosa-associated bacterial community and gene expression of tight junction proteins in the small intestine of goats. Microbiologyopen 2019, 6, 8. [Google Scholar] [CrossRef]
- Song, P.X.; Zhang, R.J.; Wang, X.X.; He, P.; Tan, L.; Ma, X. Dietary grape-seed procyanidins decreased post-weaning diarrhea by modulating intestinal permeability and suppressing oxidative stress in rats. J. Agric. Food Chem. 2011, 59, 6227–6232. [Google Scholar] [CrossRef]
- Khafipour, E.; Li, S.; Tun, H.; Derakhshani, H.; Moossavi, S.; Plaizier, J. Effects of grain feeding on microbiota in the digestive tract of cattle. Anim. Front. 2016, 6, 13–19. [Google Scholar] [CrossRef]
- Tang, Y.Q.; Shigematsu, T.; Morimura, S.; Kida, K. Microbial community analysis of mesophilic anaerobic protein degradation process using bovine serum albumin (BSA)-fed continuous cultivation. J. Biosci. Bioeng. 2005, 99, 150–164. [Google Scholar] [CrossRef]
- Tao, S.Y.; Tian, P.; Luo, Y.W.; Tian, J.; Hua, C.F.; Geng, Y.L.; Cong, R.H.; Ni, Y.D.; Zhao, R.Q. Microbiome-metabolome responses to a high-grain diet associated with the hind-gut health of goats. Front. Microbiol. 2017, 8, 1764. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, M.; Wang, S. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010, 4, 232–241. [Google Scholar] [CrossRef]
- Khafipour, E.; Li, S.; Plaizier, J.C.; Krause, D.O. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl. Environ. Microbiol. 2009, 75, 7115–7124. [Google Scholar] [CrossRef]
- Jenkins, S.; Waite, I.; Mansfield, J.; Kim, J.; Pluske, J. Relationships between diets different in fibre type and content with growth, Escherichia coli shedding, and faecal microbial diversity after weaning. Anim. Prod. Sci. 2015, 55, 1451. [Google Scholar] [CrossRef]
- Di Rienzi, S.C.; Sharon, I.; Wrighton, K.C.; Koren, O.; Hug, L.A.; Thomas, B.C.; Goodrich, J.K.; Bell, J.T.; Spector, T.D.; Banfield, J.F.; et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2013, 1, 2. [Google Scholar]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Xu, T.T.; Zhu, W.Y.; Mao, S.Y. High-grain feeding alters caecal bacterial microbiota composition and fermentation and results in caecal mucosal injury in goats. Br. J. Nutr. 2014, 112, 416–427. [Google Scholar] [CrossRef]
- Piknova, M.; Guczynska, W.; Miltko, R.; Javorsky, P.; Kasperowicz, A.; Michalowski, T.; Pristas, P. Treponema zioleckii sp nov., a novel fructanutilizing species of rumen treponemes. FEMS Microbiol. Lett. 2008, 289, 166–172. [Google Scholar] [CrossRef]
- Garcia, J.P.; Adams, V.; Beingesser, J.; Hughes, M.L.; Poon, R.; Lyras, D.; Hill, A.; McClane, B.A.; Rood, J.I.; Uzal, F.A. Epsilon toxin is essential for the virulence of Clostridium perfringens type D infection in sheep, goats, and mice. Infect. Immun. 2013, 81, 2405–2414. [Google Scholar] [CrossRef]
- Kellermayer, R.; Dowd, S.E.; Harris, R.A.; Balasa, A.; Schaible, T.D.; Wolcott, R.D. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 2011, 25, 1449–1460. [Google Scholar] [CrossRef]
- Bosshard, P.P.; Zbinden, R.; Altwegg, M. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Grampositive bacterium. Int. J. Syst. Evol. Microbiol. 2002, 52, 1263–1266. [Google Scholar]
- Wang, J.; Fan, H.; Han, Y.; Zhao, J.; Zhou, Z. Characterization of the microbial communities along the gastrointestinal tract of sheep by 454 pyroseqencing analysis. Asian-Australas. J. Anim. Sci. 2016, 1, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Bayer, E.A.; Rincon, M.T.; Lamed, R.; White, B.A. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008, 6, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Chassard, C.; Delmas, E.; Robert, C.; Lawson, P.A.; Bernalier-Donadille, A. Ruminococcus champanellensis sp. nov., a cellulose-degrading bacterium fromhuman gut microbiota. Int. J. Syst. Evol. Microbiol. 2012, 62, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, J.S.; Sonnenburg, J.L.; Elias, J.E. Monitoring host responses to the gut microbiota. ISME J. 2015, 9, 1908–1915. [Google Scholar] [CrossRef] [PubMed]
- Ussar, S.; Griffin, N.W.; Bezy, O. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab. 2015, 22, 516–530. [Google Scholar] [CrossRef]
- Konishi, H.; Fujiya, M.; Kohgo, Y. Host–microbe interactions via membrane transport systems. Environ. Microbiol. 2015, 17, 931–937. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469. [Google Scholar] [CrossRef]
- Shibata, N.; Kunisawa, J.; Kiyono, H. Dietary and microbial metabolites in the regulation of host immunity. Front. Microbiol. 2017, 8, 2171. [Google Scholar] [CrossRef]
- Harrison, E.H.; Hussain, M.M. Mechanisms involved in the intestinal digestion and absorption of dietary vitamin A. J. Nutr. 2001, 131, 1405. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Steele, M.A.; Croom, J.; Kahler, M.; AlZahal, O.; Hook, S.E.; Plaizier, K.; McBride, B.W. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Xu, T.T.; Liu, Y.J.; Zhu, W.Y.; Mao, S.Y. A high-grain diet causes massive disruption of ruminal epithelial tight junctions in goats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R232–R241. [Google Scholar] [CrossRef] [PubMed]
- Zebeli, Q.; Dunn, S.M.; Ametaj, B.N. Perturbations of plasma metabolites correlated with the rise of rumen endotoxin in dairy cows fed diets rich in easily degradable carbohydrates. J. Dairy Sci. 2011, 94, 2374–2382. [Google Scholar] [CrossRef]
- Rashid, S.; Irshadullah, M. Evaluation of antioxidant and oxidant status of goats (Capra aegagrus hircus) naturally infected with Haemonchus contortus. J. Helminthol. 2019, 94, e36. [Google Scholar] [CrossRef] [PubMed]
- Selwood, T.; Jaffe, E.K. Dynamic dissociating homo-oligomers and the control of protein function. Arch. Biochem. Biophys. 2012, 519, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Luo, Y.; Wang, H.; Liu, S.; Shen, Y.; Wang, M. Effects of Glucose and Starch on Lactate Production by Newly Isolated Streptococcus bovis S1 from Saanen Goats. Appl. Environ. Microbiol. 2016, 19, 82. [Google Scholar] [CrossRef]
- Wang, X.B.; Ogawa, T.; Suda, S.; Taniguchi, K.; Uike, H.; Kumagai, H.; Mitani, K. Effects of nutritional level on digestive enzyme activities in the pancreas and small intestine of calves slaughtered at same body weight. Asian-Australas. J. Anim. Sci. 1998, 11, 375–380. [Google Scholar] [CrossRef]
- Owsley, W.F.; Orr, D.E.; Tribble, L.F. Effects of age and diet on the development of the pancreas and the synthesis and secretion of pancreatic enzymes in the young pig. J. Anim. Sci. 1986, 63, 497–504. [Google Scholar] [CrossRef]
- Liu, J.H.; Xu, T.T.; Zhu, W.Y.; Mao, S.Y. A high-grain diet alters the omasal epithelial structure and expression of tight junction proteins in a goat model. Vet. J. 2014, 1, 95–100. [Google Scholar] [CrossRef]




| Items | NC 1 (n = 8) | HC 2 (n = 8) |
|---|---|---|
| Ingredients composition (%) | ||
| Rice straw | 45.0 | 10.0 |
| Rice with shell | 33.2 | 54.3 |
| Soybean meal | 9.60 | 15.7 |
| Wheat bran | 6.00 | 9.80 |
| Fat powder | 3.20 | 5.20 |
| Calcium carbonate | 0.500 | 0.800 |
| Calcium bicarbonate | 1.10 | 1.80 |
| Sodium chloride | 0.600 | 1.00 |
| Premix 3 | 1.00 | 1.40 |
| Nutrient levels 4, % of DM (dry matter) | ||
| Crude protein | 13.5 | 17.6 |
| Crude ash | 9.34 | 9.12 |
| Crude fat | 4.18 | 6.01 |
| Neutral detergent fiber | 49.8 | 38.4 |
| Acid detergent fiber | 36.5 | 9.51 |
| Items | Intra-Assay Variation | Inter-Assay Variation |
|---|---|---|
| LACT 1 | cv %: ≤2% | cv %: ≤3% |
| LDH 2 | cv %: ≤3% | cv %: ≤5% |
| ALT 3 | cv %: ≤3% | cv %: ≤4% |
| AST 4 | cv %: ≤3% | cv %: ≤4% |
| ALP 5 | cv %: ≤2% | cv %: ≤3% |
| AMY 6 | cv %: ≤2% | cv %: ≤3% |
| Phylum | NC 1 (n = 6) | HC 2 (n = 6) | p-Value |
|---|---|---|---|
| Actinobacteria | 2.88 ± 2.21 | 2.37± 1.06 | 0.624 |
| Bacteroidetes | 0.90 ± 2.08 | 1.07 ± 0.80 | 0.853 |
| Chloroflexi | 1.08 ± 1.21 | 0.02 ± 0.02 | 0.058 |
| Cyanobacteria | 0.83 ± 0.66 | 0.25 ± 0.42 | 0.090 |
| Elusimicrobia | 0.91 ± 0.93 | 0.96 ± 2.16 | 0.954 |
| Euryarchaeota | 0.64 ± 0.46 | 0.23 ± 0.10 | 0.055 |
| Firmicutes | 79.5 ± 10.3 | 80.2 ± 18.6 | 0.945 |
| Lentisphaerae | 1.36 ± 0.92 | 1.13 ± 1.25 | 0.721 |
| Proteobacteria | 0.83 ± 0.82 | 1.12 ± 1.23 | 0.631 |
| Saccharibacteria | 2.38 ± 1.64 | 1.16 ± 1.38 | 0.191 |
| Tenericutes | 5.92 ± 4.20 | 10.5 ± 8.4 | 0.570 |
| Verrucomicrobia | 2.50 ± 2.00 | 0.86 ± 0.66 | 0.220 |
| Classification Levels of Bacteria | Abundance (%) | p-Value | |||
|---|---|---|---|---|---|
| Phylum | Family | Genus | NC 1 (n = 6) | HC 2 (n = 6) | |
| Actinobacteria | Coriobacteriaceae | Senegalimassilia | 1.39 ± 1.28 | 0.43 ± 0.21 | 0.099 |
| Elusimicrobia | Elusimicrobiaceae | Elusimicrobium | 0.91 ± 0.93 | 0.96 ± 2.16 | 0.956 |
| Firmicutes | Christensenellaceae | Christensenellaceae_R-7_group | 18.2 ± 12.1 | 12.3 ± 5.11 | 0.149 |
| Clostridiaceae_1 | Clostridium_sensu_stricto_1 | 0.26 ± 0.11 | 1.13 ± 0.62 | 0.022 | |
| Erysipelotrichaceae | Turicibacter | 0.82 ± 0.62 | 1.19 ± 1.73 | 0.641 | |
| Family_XIII | Eubacterium_nodatum_group | 0.53 ± 0.28 | 1.26 ± 0.63 | 0.026 | |
| Family_XIII | Family_XIII_AD3011_group | 2.21 ± 0.99 | 2.31 ± 0.67 | 0.846 | |
| Family_XIII | Mogibacterium | 2.59 ± 2.26 | 2.72 ± 1.35 | 0.903 | |
| Lachnospiraceae | Acetitomaculum | 1.12 ± 0.89 | 1.57 ± 1.56 | 0.556 | |
| Lachnospiraceae | Eubacterium_ventriosum_group | 0.23 ± 0.45 | 1.53 ± 2.35 | 0.236 | |
| Lachnospiraceae | Lachnospiraceae_NK3A20_group | 2.20 ± 1.59 | 2.37 ± 1.43 | 0.850 | |
| Lachnospiraceae | Ruminococcus_gauvreauii_group | 0.28 ± 0.28 | 1.42 ± 0.91 | 0.026 | |
| Peptostreptococcaceae | Peptoclostridium | 5.71 ± 4.44 | 1.40 ± 2.27 | 0.061 | |
| Peptostreptococcaceae | Romboutsia | 7.71 ± 4.53 | 5.00 ± 6.05 | 0.402 | |
| Ruminococcaceae | Anaerotruncus | 1.79 ± 0.99 | 0.23 ± 0.10 | 0.012 | |
| Ruminococcaceae | Eubacterium_coprostanoligenes_group | 1.07 ± 0.59 | 4.51 ± 3.38 | 0.034 | |
| Ruminococcaceae | Ruminococcaceae_NK4A214_group | 6.06 ± 3.24 | 7.79 ± 4.68 | 0.472 | |
| Ruminococcaceae | Ruminococcaceae_UCG-001 | 0.20 ± 0.28 | 1.24 ± 1.92 | 0.247 | |
| Ruminococcaceae | Ruminococcaceae_UCG-014 | 3.32 ± 3.41 | 9.36 ± 9.22 | 0.180 | |
| Ruminococcaceae | Ruminococcus 1 | 0.18 ± 0.36 | 1.00 ± 0.72 | 0.031 | |
| Ruminococcaceae | Ruminococcus_1 | 0.30 ± 0.23 | 0.99 ± 0.76 | 0.080 | |
| Ruminococcaceae | Ruminococcus_2 | 1.77 ± 1.62 | 5.24 ± 4.82 | 0.144 | |
| Ruminococcaceae | Saccharofermentans | 9.76 ± 2.15 | 5.70 ± 7.06 | 0.495 | |
| Saccharibacteria | Unknown | Candidatus_Saccharimonas | 2.38 ± 1.64 | 1.16 ± 1.38 | 0.191 |
| Tenericutes | Mycoplasmataceae | Mycoplasma | 3.92 ± 4.79 | 8.65 ± 18.8 | 0.564 |
| Unidentified | 14.6 ± 6.96 | 9.32 ± 2.45 | 0.127 | ||
| Level 2 | Level 3 | Pathway ID | NC 1 (n = 6) | HC 2 (n = 6) | p-Value |
|---|---|---|---|---|---|
| Amino acid metabolism | Cysteine and methionine metabolism | ko00270 | 1.01 ± 0.03 | 0.97 ± 0.06 | 0.055 |
| Histidine metabolism | ko00340 | 0.62 ± 0.02 | 0.65 ± 0.01 | 0.010 | |
| Valine, leucine and isoleucine biosynthesis | ko00290 | 0.75 ± 0.02 | 0.79 ± 0.04 | 0.050 | |
| Valine, leucine and isoleucine degradation | ko00280 | 0.22 ± 0.03 | 0.19 ± 0.02 | 0.037 | |
| Biosynthesis of other secondary metabolites | Novobiocin biosynthesis | ko00401 | 0.16 ± 0.005 | 0.14 ± 0.01 | 0.004 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | ko00960 | 0.14 ± 0.005 | 0.13 ± 0.007 | 0.004 | |
| Carbohydrate metabolism | Butanoate metabolism | ko00650 | 0.77 ± 0.02 | 0.70 ± 0.06 | 0.010 |
| Galactose metabolism | ko00052 | 0.63 ± 0.03 | 0.68 ± 0.05 | 0.055 | |
| Pentose and glucuronate interconversions | ko00040 | 0.48 ± 0.03 | 0.53 ± 0.01 | 0.004 | |
| Pentose phosphate pathway | ko00030 | 0.86 ± 0.03 | 0.95 ± 0.03 | 0.004 | |
| Pyruvate metabolism | ko00620 | 1.12 ± 0.03 | 1.16 ± 0.04 | 0.037 | |
| Starch and sucrose metabolism | ko00500 | 0.94 ± 0.03 | 1.02 ± 0.04 | 0.010 | |
| Cell motility | Bacterial chemotaxis | ko02030 | 0.70 ± 0.12 | 0.58 ± 0.02 | 0.006 |
| Flagellar assembly | ko02040 | 0.70 ± 0.11 | 0.56 ± 0.07 | 0.037 | |
| Glycan biosynthesis and metabolism | Other glycan degradation | ko00511 | 0.12 ± 0.01 | 0.14 ± 0.01 | 0.025 |
| Lipid metabolism | Fatty acid biosynthesis | ko00061 | 0.52 ± 0.02 | 0.55 ± 0.03 | 0.055 |
| Glycerolipid metabolism | ko00561 | 0.42 ± 0.02 | 0.46 ± 0.02 | 0.016 | |
| Metabolism of cofactors and vitamins | Nicotinate and nicotinamide metabolism | ko00760 | 0.41 ± 0.01 | 0.45 ± 0.02 | 0.004 |
| Riboflavin metabolism | ko00740 | 0.21 ± 0.02 | 0.19 ± 0.02 | 0.025 | |
| Vitamin B6 metabolism | ko00750 | 0.17 ± 0.01 | 0.20 ± 0.02 | 0.025 | |
| Metabolism of other amino acids | beta-Alanine metabolism | ko00410 | 0.16 ± 0.02 | 0.14 ± 0.01 | 0.025 |
| Metabolism of terpenoids and polyketides | Biosynthesis of ansamycins | ko01051 | 0.12 ± 0.003 | 0.13 ± 0.003 | 0.004 |
| Tetracycline biosynthesis | ko00253 | 0.18 ± 0.009 | 0.20 ± 0.01 | 0.010 | |
| Signal transduction | Two-component system | ko02020 | 1.52 ± 0.11 | 1.37 ± 0.07 | 0.010 |
| Xenobiotics biodegradation and metabolism | Nitrotoluene degradation | ko00633 | 0.13 ± 0.004 | 0.09 ± 0.02 | 0.004 |
| Polycyclic aromatic hydrocarbon degradation | ko00624 | 0.09 ± 0.005 | 0.11 ± 0.007 | 0.016 |
| Items | NC 1 (n = 6) | HC 2 (n = 6) | p Value |
|---|---|---|---|
| LPS 3 (EU/mL) | 0.41 ± 0.04 | 0.36 ± 0.05 | 0.136 |
| LACT 4 (mmol/L) | 0.14 ± 0.05 | 0.20 ± 0.14 | 0.343 |
| LDH 5 (U/L) | 4.83 ± 2.64 | ND 6 | 0.006 |
| ALT 7 (U/L) | 2.00 ± 0.58 | 1.55 ± 0.56 | 0.206 |
| AST 8 (U/L) | 7.05 ± 4.86 | 6.17 ± 4.18 | 0.743 |
| ALP 9 (U/mL) | 5.95 ± 2.26 | 8.76 ± 0.83 | 0.017 |
| AMY 10 (U/L) | 195 ± 18 | 272 ± 53 | 0.014 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Ren, A.; Zheng, M.; Jiao, J.; Yan, Q.; Zhou, C.; Tan, Z. Diet with a High Proportion of Rice Alters Profiles and Potential Function of Digesta-Associated Microbiota in the Ileum of Goats. Animals 2020, 10, 1261. https://doi.org/10.3390/ani10081261
Wang K, Ren A, Zheng M, Jiao J, Yan Q, Zhou C, Tan Z. Diet with a High Proportion of Rice Alters Profiles and Potential Function of Digesta-Associated Microbiota in the Ileum of Goats. Animals. 2020; 10(8):1261. https://doi.org/10.3390/ani10081261
Chicago/Turabian StyleWang, Kaijun, Ao Ren, Mengli Zheng, Jinzhen Jiao, Qiongxian Yan, Chuanshe Zhou, and Zhiliang Tan. 2020. "Diet with a High Proportion of Rice Alters Profiles and Potential Function of Digesta-Associated Microbiota in the Ileum of Goats" Animals 10, no. 8: 1261. https://doi.org/10.3390/ani10081261
APA StyleWang, K., Ren, A., Zheng, M., Jiao, J., Yan, Q., Zhou, C., & Tan, Z. (2020). Diet with a High Proportion of Rice Alters Profiles and Potential Function of Digesta-Associated Microbiota in the Ileum of Goats. Animals, 10(8), 1261. https://doi.org/10.3390/ani10081261






