Comparative Transcriptomic Analysis of the Pituitary Gland between Cattle Breeds Differing in Growth: Yunling Cattle and Leiqiong Cattle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals and Sample Collection
2.3. Quantification of Growth-Related Hormones
2.4. RNA Extraction and Sequencing
2.5. RNA Seq Data Assembly and Functional Assignment
2.6. Verification of Sequencing Data by RT-qPCR
2.7. Functional Gene Annotation
2.8. PPI Network Construction
3. Results
3.1. Growth-Related Hormones’ Expression Analysis
3.2. Summary of Mapping Statistics
3.3. Characteristics of Expression Profile Data
3.4. Validation of Samples’ Reproduction and RNA-seq Data’s Accuracy
3.5. Identification of DEGs
3.6. Enrichment Analysis of DEGs
3.7. PPI Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hocquette, J.; Gondret, F.; Baéza, E.; Médale, F.; Jurie, C.; Pethick, D.W. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2009, 4, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, F.; He, C.; Zhang, L.; Tan, D.; Reiter, R.J.; Xu, J.; Ji, P.; Liu, G.J. Beneficial effects of melatonin on bovine oocytes maturation: A mechanistic approach. J. Pineal Res. 2014, 57, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.W.; Ellsworth, B.S.; Millan, M.I.P.; Gergics, P.; Schade, V.; Foyouzi, N.; Brinkmeier, M.L.; Mortensen, A.H.; Camper, S.A. Pituitary Gland Development and Disease. In Current Topics in Developmental Biology; Elsevier BV: Amsterdam, The Netherlands, 2013; Volume 106, pp. 1–47. [Google Scholar]
- Lucy, M. Functional Differences in the Growth Hormone and Insulin-like Growth Factor Axis in Cattle and Pigs: Implications for Post-partum Nutrition and Reproduction. Reprod. Domest. Anim. 2008, 43, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Spicer, L.J.; Aad, P. Insulin-Like Growth Factor (IGF) 2 Stimulates Steroidogenesis and Mitosis of Bovine Granulosa Cells Through the IGF1 Receptor: Role of Follicle-Stimulating Hormone and IGF2 Receptor1. Biol. Reprod. 2007, 77, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Gkourogianni, A.; Andrade, A.C.; Jonsson, B.; Segerlund, E.; Werner-Sperker, A.; Horemuzova, E.; Dahlgren, J.; Burstedt, M.; Nilsson, O. Pre- and postnatal growth failure with microcephaly due to two novel heterozygous IGF1R mutations and response to growth hormone treatment. Acta Paediatr. 2020, 00, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chagas, L.; Bass, J.; Blache, D.; Burke, C.; Kay, J.; Lindsay, D.; Lucy, M.; Martin, G.B.; Meier, S.; Rhodes, F.; et al. Invited Review: New Perspectives on the Roles of Nutrition and Metabolic Priorities in the Subfertility of High-Producing Dairy Cows. J. Dairy Sci. 2007, 90, 4022–4032. [Google Scholar] [CrossRef]
- Fonseca, P.A.D.S.; Id-Lahoucine, S.; Reverter, A.; Medrano, J.F.; Fortes, M.R.S.; Casellas, J.; Miglior, F.; Brito, L.; Carvalho, M.R.S.; Schenkel, F.S.; et al. Combining multi-OMICs information to identify key-regulator genes for pleiotropic effect on fertility and production traits in beef cattle. PLoS ONE 2018, 13, e0205295. [Google Scholar] [CrossRef]
- Pal, R.P.; Mani, V.; Tripathi, D.; Kumar, R.; KewalRamani, N.J. Influence of Feeding Inorganic Vanadium on Growth Performance, Endocrine Variables and Biomarkers of Bone Health in Crossbred Calves. Biol. Trace Elem. Res. 2017, 182, 248–256. [Google Scholar] [CrossRef]
- Seabury, C.; Oldeschulte, D.; Saatchi, M.; Beever, J.E.; Decker, J.E.; Halley, Y.A.; Bhattarai, E.K.; Molaei, M.; Freetly, H.C.; Hansen, S.L.; et al. Genome-wide association study for feed efficiency and growth traits in U.S. beef cattle. BMC Genom. 2017, 18, 386. [Google Scholar] [CrossRef]
- Steele, M.; Penner, G.B.; Chaucheyras-Durand, F.; Guan, L.L. Development and physiology of the rumen and the lower gut: Targets for improving gut health. J. Dairy Sci. 2016, 99, 4955–4966. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Reverter, A.; Venus, B.; Islas-Trejo, A.; Porto-Neto, L.R.; Lehnert, S.A.; Medrano, J.F.; Moore, S.S.; Fortes, M.R.S.; Cánovas, A. Global differential gene expression in the pituitary gland and the ovaries of pre- and postpubertal Brahman heifers1. J. Anim. Sci. 2017, 95, 599–615. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xia, X.; Qu, K.; Li, F.; Jia, P.; Chen, Q.; Chen, N.; Zhang, J.; Chen, H.; Huang, B.; Lei, C. Abundant Genetic Diversity of Yunling Cattle Based on Mitochondrial Genome. Animals 2019, 9, 641. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-P.; Geng, R.-Q.; Chang, H. Mitochondrial DNA Diversity and Origin of Chinese Leiqiong Cattle. J. Anim. Vet. Adv. 2009, 8, 1312–1315. [Google Scholar]
- Zhang, F.; Qu, K.; Chen, N.; Hanif, Q.; Jia, Y.; Huang, Y.-Z.; Dang, R.; Zhang, J.; Lan, X.; Chen, H.; et al. Genome-Wide SNPs and InDels Characteristics of Three Chinese Cattle Breeds. Animals 2019, 9, 596. [Google Scholar] [CrossRef]
- He, R.; Gu, X.; Lai, W.; Peng, X.; Yang, G.-Y. Transcriptome-microRNA analysis of Sarcoptes scabiei and host immune response. PLoS ONE 2017, 12, e0177733. [Google Scholar] [CrossRef]
- Huang, G.; Zhao, G.; Xia, J.; Wei, Y.; Chen, F.; Chen, J.; Shi, J. FGF2 and FAM201A affect the development of osteonecrosis of the femoral head after femoral neck fracture. Gene 2018, 652, 39–47. [Google Scholar] [CrossRef]
- Kroll, K.W.; Mokaram, N.E.; Pelletier, A.R.; Frankhouser, D.E.; Westphal, M.S.; Stump, P.A.; Stump, C.L.; Bundschuh, R.; Blachly, J.S.; Yan, P.S. Quality Control for RNA-Seq (QuaCRS): An Integrated Quality Control Pipeline. Cancer Inform. 2014, 13, 7–14. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 31. [Google Scholar] [CrossRef]
- Thornton, B.; Basu, C. Rapid and Simple Method of qPCR Primer Design. Adv. Struct. Saf. Stud. 2015, 1275, 173–179. [Google Scholar]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Leung, F.K.C.; Stuart, M.C.A.; Kajitani, T.; Fukushima, T.; Van Der Giessen, E.; Feringa, B.L. Artificial muscle-like function from hierarchical supramolecular assembly of photoresponsive molecular motors. Nat. Chem. 2017, 10, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Mullur, R.; Liu, Y.-Y.; Brent, G.A. Thyroid hormone regulation of metabolism. Physiol. Rev. 2014, 94, 355–382. [Google Scholar] [CrossRef]
- Tsigos, C.; Chrousos, G.P. Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and stress. J. Psychosom. Res. 2002, 53, 865–871. [Google Scholar] [CrossRef]
- Gholizadeh, M.; Fayazi, J.; Asgari, Y.; Zali, H.; Kaderali, L. Reconstruction and Analysis of Cattle Metabolic Networks in Normal and Acidosis Rumen Tissue. Animals 2020, 10, 469. [Google Scholar] [CrossRef]
- Jiang, R.; Li, H.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Transcriptome profiling of lncRNA related to fat tissues of Qinchuan cattle. Gene 2020, 742, 144587. [Google Scholar] [CrossRef]
- Yan, X.-M.; Zhang, Z.; Meng, Y.; Li, H.-B.; Gao, L.; Luo, D.; Jiang, H.; Gao, Y.; Yuan, B.; Zhang, J.-B. Genome-wide identification and analysis of circular RNAs differentially expressed in the longissimus dorsi between Kazakh cattle and Xinjiang brown cattle. PeerJ 2020, 8, e8646. [Google Scholar]
- Giustina, A.; Mazziotti, G.; Canalis, E. Growth hormone, insulin-like growth factors, and the skeleton. Endocr. Rev. 2008, 29, 535–559. [Google Scholar] [CrossRef]
- Webb, E.C.; Casey, N. Physiological limits to growth and the related effects on meat quality. Livest. Sci. 2010, 130, 33–40. [Google Scholar] [CrossRef]
- Huszenicza, G.; Kulcsar, M.; Kóródi, P.; Bartyik, J.; Rudas, P.; Ribiczei-Szabo, P.; Nikolić-Judith, A.; Šamanc, H.; Ivanov, I.; Gvozdić, D. Adrenocortical and thyroid function, hormone and metabolite profiles and the onset of ovarian cyclicity in dairy cows suffering from various forms of ketosis. Acta Vet. 2006, 56, 25–36. [Google Scholar] [CrossRef]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Hornick, J.-L.; Van Eenaeme, C.; Gérard, O.; Dufrasne, I.; Istasse, L. Mechanisms of reduced and compensatory growth. Domest. Anim. Endocrinol. 2000, 19, 121–132. [Google Scholar] [CrossRef]
- Wang, P.; Drackley, J.; Stamey-Lanier, J.; Keisler, D.; Loor, J. Effects of level of nutrient intake and age on mammalian target of rapamycin, insulin, and insulin-like growth factor-1 gene network expression in skeletal muscle of young Holstein calves. J. Dairy Sci. 2014, 97, 383–391. [Google Scholar] [CrossRef]
- Qin, L.; Xiang, Y.; Song, Z.; Jing, R.; Hu, C.; Howard, S.T. Retraction notice to “Erythropoietin as a possible mechanism for the effects of intermittent hypoxia on bodyweight, serum glucose and leptin in mice”. Regul. Pept. 2010, 165, 168–173. [Google Scholar] [CrossRef]
- Sauerwein, H.; Bendixen, E.; Restelli, L.; Ceciliani, F. The adipose tissue in farm animals: A proteomic approach. Curr. Protein Pept. Sci. 2014, 15, 146–155. [Google Scholar] [CrossRef]
- Grace, F.M.; Scultorpe, N.; Solomon, A.; Bouloux, P.; Lewis, M. Androgens Affect Myogenesis In Vitro in Association with Increased Local Igf-1 Expression. Med. Sci. Sports Exerc. 2011, 43, 413. [Google Scholar] [CrossRef]
- Xu, W.; Morford, J.; Mauvais-Jarvis, F. Emerging role of testosterone in pancreatic β cell function and insulin secretion. J. Endocrinol. 2019, 240, R97–R105. [Google Scholar] [CrossRef]
- Cristina, R.; Hanganu, F.; Dumitrescu, E.; Muselin, F.; Butnariu, M.; Constantin, A.; Chiurciu, V. The Impact of Exogenic Testosterone and Nortestosterone-Decanoate Toxicological Evaluation Using a Rat Model. PLoS ONE 2014, 9, e109219. [Google Scholar] [CrossRef]
- Sun, X.; Li, M.; Sun, Y.; Cai, H.; Lan, X.; Huang, Y.; Bai, Y.; Qi, X.; Chen, H. The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2016, 1863, 2835–2845. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Li, D.; Wang, G.; Zhang, C.; Zhang, M.; Zhang, W.; Li, S. Three intronic lncRNAs with monoallelic expression derived from the MEG8 gene in cattle. Anim. Genet. 2016, 48, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lu, X.; Xia, H.; Zhang, C.; Wang, X.; Chen, Z.; Zhang, H.; Qu, K.; Huang, B.; Moore, S.; et al. In-depth characterization of the pituitary transcriptome in Simmental and Chinese native cattle. Domest. Anim. Endocrinol. 2019, 66, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The Transcriptional Landscape of the Mammalian Genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Inoki, K.; Li, Y.; Xu, T.; Guan, K.-L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003, 17, 1829–1834. [Google Scholar] [CrossRef]
- Yaba, A.; Demir, N. The mechanism of mTOR (mammalian target of rapamycin) in a mouse model of polycystic ovary syndrome (PCOS). J. Ovarian Res. 2012, 5, 38. [Google Scholar] [CrossRef]
- Huang, E.J.; Reichardt, L.F. Trk Receptors: Roles in Neuronal Signal Transduction. Annu. Rev. Biochem. 2003, 72, 609–642. [Google Scholar] [CrossRef]
- Damonte, V.M.; Rodríguez, S.S.; Raingo, J.; Valentina, M.D.; Susana, R.S.; Jesica, R. Growth hormone secretagogue receptor constitutive activity impairs voltage-gated calcium channel-dependent inhibitory neurotransmission in hippocampal neurons. J. Physiol. 2018, 596, 5415–5428. [Google Scholar] [CrossRef]
- Dalrymple, B.P.; Guo, B. Triennial Growth and Development Symposium: Intramuscular fat deposition in ruminants and pigs: A transcriptomics perspective1. J. Anim. Sci. 2017, 95, 2272–2283. [Google Scholar] [CrossRef]
- Sinclair, K.D.; A Lunn, L.; Kwong, W.Y.; Wonnacott, K.; Linforth, R.; Craigon, J. Amino acid and fatty acid composition of follicular fluid as predictors of in-vitro embryo development. Reprod. Biomed. Online 2008, 16, 859–868. [Google Scholar] [CrossRef]
- Divya, B.K.; Mohindra, V.; Singh, R.K.; Yadav, P.; Masih, P.; Jena, J.K. Muscle transcriptome resource for growth, lipid metabolism and immune system in Hilsa shad, Tenualosa ilisha. Genes Genom. 2018, 41, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Liang, G.; Zhu, X.; Tan, Y.; Xu, J.; Wu, H.; Mao, H.; Zhang, Y.; Chen, J.; Rao, Y.; et al. Gene Expression Profiling in Ovaries and Association Analyses Reveal HEP21 as a Candidate Gene for Sexual Maturity in Chickens. Animals 2020, 10, 181. [Google Scholar] [CrossRef] [PubMed]
- Komenoi, S.; Suzuki, Y.; Asami, M.; Murakami, C.; Hoshino, F.; Chiba, S.; Takahashi, D.; Kado, S.; Sakane, F. Microarray analysis of gene expression in the diacylglycerol kinase η knockout mouse brain. Biochem. Biophys. Rep. 2019, 19, 100660. [Google Scholar] [CrossRef]
- Yang, F.; Chen, F.; Li, L.; Yan, L.; Badri, T.; Lv, C.; Yu, D.; Zhang, M.; Jang, X.; Li, J.; et al. Three Novel Players: PTK2B, SYK, and TNFRSF21 Were Identified to Be Involved in the Regulation of Bovine Mastitis Susceptibility via GWAS and Post-transcriptional Analysis. Front. Immunol. 2019, 10, 1579. [Google Scholar] [CrossRef]
- Vaughan, O.R.; Sferruzzi-Perri, A.N.; Fowden, A.L. Maternal corticosterone regulates nutrient allocation to fetal growth in mice. J. Physiol. 2012, 590, 5529–5540. [Google Scholar] [CrossRef]
- Chan, K.; Busque, S.M.; Sailer, M.; Stoeger, C.; Broer, S.; Daniel, H.; Rubio-Aliaga, I.; Wagner, C.A. Loss of function mutation of the Slc38a3 glutamine transporter reveals its critical role for amino acid metabolism in the liver, brain, and kidney. Pflügers Arch.-Eur. J. Physiol. 2015, 468, 213–227. [Google Scholar] [CrossRef]
- Kolch, W. Meaningful relationships: The regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000, 351, 289. [Google Scholar] [CrossRef]
- Widmann, P.; Reverter, A.; Fortes, M.R.S.; Weikard, R.; Suhre, K.; Hammon, H.M.; Albrecht, E.; Kühn, C. A systems biology approach using metabolomic data reveals genes and pathways interacting to modulate divergent growth in cattle. BMC Genom. 2013, 14, 798. [Google Scholar] [CrossRef]
- Wettschureck, N.; Moers, A.; Wallenwein, B.; Parlow, A.F.; Maser-Gluth, C.; Offermanns, S. Loss of Gq/11 Family G Proteins in the Nervous System Causes Pituitary Somatotroph Hypoplasia and Dwarfism in Mice. Mol. Cell. Biol. 2005, 25, 1942–1948. [Google Scholar] [CrossRef]
- Zhai, M.; Zhao, Z.; Yang, M.; Liang, Y.; Liang, H.; Xie, Y.; Han, J. The effect of GNAQ methylation on GnRH secretion in sheep hypothalamic neurons. J. Cell. Biochem. 2019, 120, 19396–19405. [Google Scholar] [CrossRef] [PubMed]
- Arnaud-Lopez, L.; Usala, G.; Ceresini, G.; Mitchell, B.D.; Pilia, M.G.; Piras, M.G.; Sestu, N.; Maschio, A.; Busonero, F.; Albai, G.; et al. Phosphodiesterase 8B Gene Variants Are Associated with Serum TSH Levels and Thyroid Function. Am. J. Hum. Genet. 2008, 82, 1270–1280. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Hiraide, T.; Yamoto, K.; Nakashima, M.; Kawai, T.; Yanagi, K.; Ogata, T.; Saitsu, H. Exome reports A de novo GNB2 variant associated with global developmental delay, intellectual disability, and dysmorphic features. Eur. J. Med. Genet. 2020, 63, 103804. [Google Scholar] [CrossRef] [PubMed]
- Gelsomino, L.; Gu, G.; Rechoum, Y.; Beyer, A.R.; Pejerrey, S.M.; Tsimelzon, A.; Wang, T.; Huffman, K.; Ludlow, A.; Andò, S.; et al. ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res. Treat. 2016, 157, 253–265. [Google Scholar] [CrossRef]
- Sárközy, M.; Szűcs, G.; Pipicz, M.; Zvara, Á.; Éder, K.; Fekete, V.; Szűcs, C.; Bárkányi, J.; Csonka, C.; Puskás, L.G.; et al. The effect of a preparation of minerals, vitamins and trace elements on the cardiac gene expression pattern in male diabetic rats. Cardiovasc. Diabetol. 2015, 14, 85. [Google Scholar] [CrossRef]
- Ortiga-Carvalho, T.M.; Sidhaye, A.R.; Wondisford, F.E. Thyroid hormone receptors and resistance to thyroid hormone disorders. Nat. Rev. Endocrinol. 2014, 10, 582–591. [Google Scholar] [CrossRef]
- Jones, I.; Srinivas, M.; Ng, L.; Forrest, U. The Thyroid Hormone ReceptorβGene: Structure and Functions in the Brain and Sensory Systems. Thyroid 2003, 13, 1057–1068. [Google Scholar] [CrossRef]
- Payne, S.H. The utility of protein and mRNA correlation. Trends Biochem. Sci. 2014, 40, 1–3. [Google Scholar] [CrossRef]
- Rochus, C.M.; Tortereau, F.; Plisson-Petit, F.; Restoux, G.; Moreno-Romieux, C.; Tosser-Klopp, G.; Servin, B. Revealing the selection history of adaptive loci using genome-wide scans for selection: An example from domestic sheep. BMC Genom. 2018, 19, 71. [Google Scholar] [CrossRef]
- Lim, D.; Gondro, C.; Park, H.S.; Cho, Y.M.; Chai, H.H.; Seong, H.H.; Yang, B.S.; Hong, S.K.; Chang, W.K.; Lee, S.H. Identification of Recently Selected Mutations Driven by Artificial Selection in Hanwoo (Korean Cattle). Asian-Australas. J. Anim. Sci. 2013, 26, 603–608. [Google Scholar] [CrossRef]
- Hayes, B.J.; Chamberlain, A.J.; MacEachern, S.; Savin, K.; McPartlan, H.; MacLeod, I.; Sethuraman, L.; Goddard, M. A genome map of divergent artificial selection betweenBos taurusdairy cattle andBos taurusbeef cattle. Anim. Genet. 2009, 40, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Bolormaa, S.; Pryce, J.; Kemper, K.E.; Savin, K.; Hayes, B.J.; Barendse, W.; Zhang, Y.; Reich, C.M.; Mason, B.A.; Bunch, R.J.; et al. Accuracy of prediction of genomic breeding values for residual feed intake and carcass and meat quality traits in Bos taurus, Bos indicus, and composite beef cattle1. J. Anim. Sci. 2013, 91, 3088–3104. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, R.C.; Alves, B.R.C.; Prezotto, L.D.; Thorson, J.F.; O Tedeschi, L.; Keisler, D.; Amstalden, M.; Williams, G. Reciprocal changes in leptin and NPY during nutritional acceleration of puberty in heifers. J. Endocrinol. 2014, 223, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Fortes, M.R.S.; Porto-Neto, L.; Kelly, M.; Venus, B.; Kidd, L.; Rego, J.P.A.D.; Edwards, S.; Boe-Hansen, G.; Piper, E.; et al. Candidate Gene Expression in Bos indicus Ovarian Tissues: Prepubertal and Postpubertal Heifers in Diestrus. Front. Vet. Sci. 2016, 3. [Google Scholar] [CrossRef]

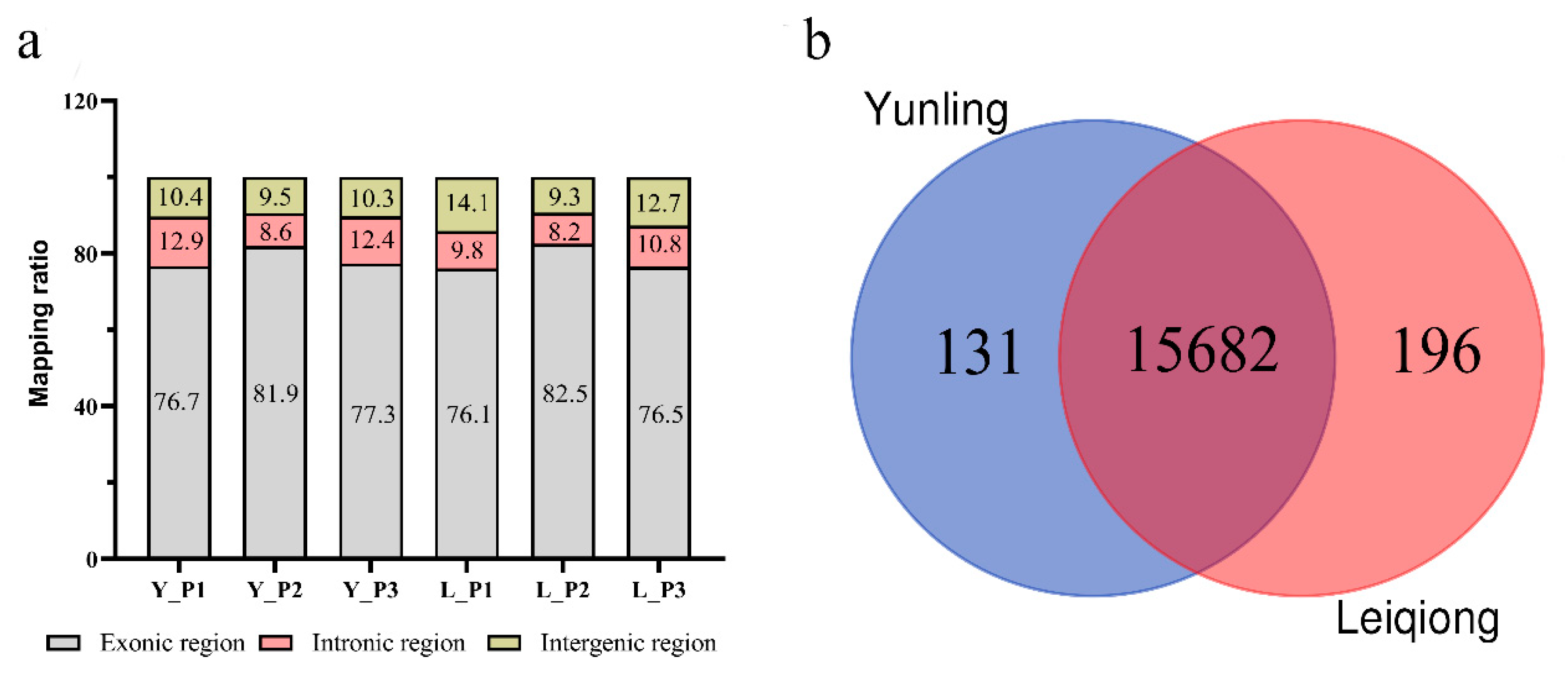
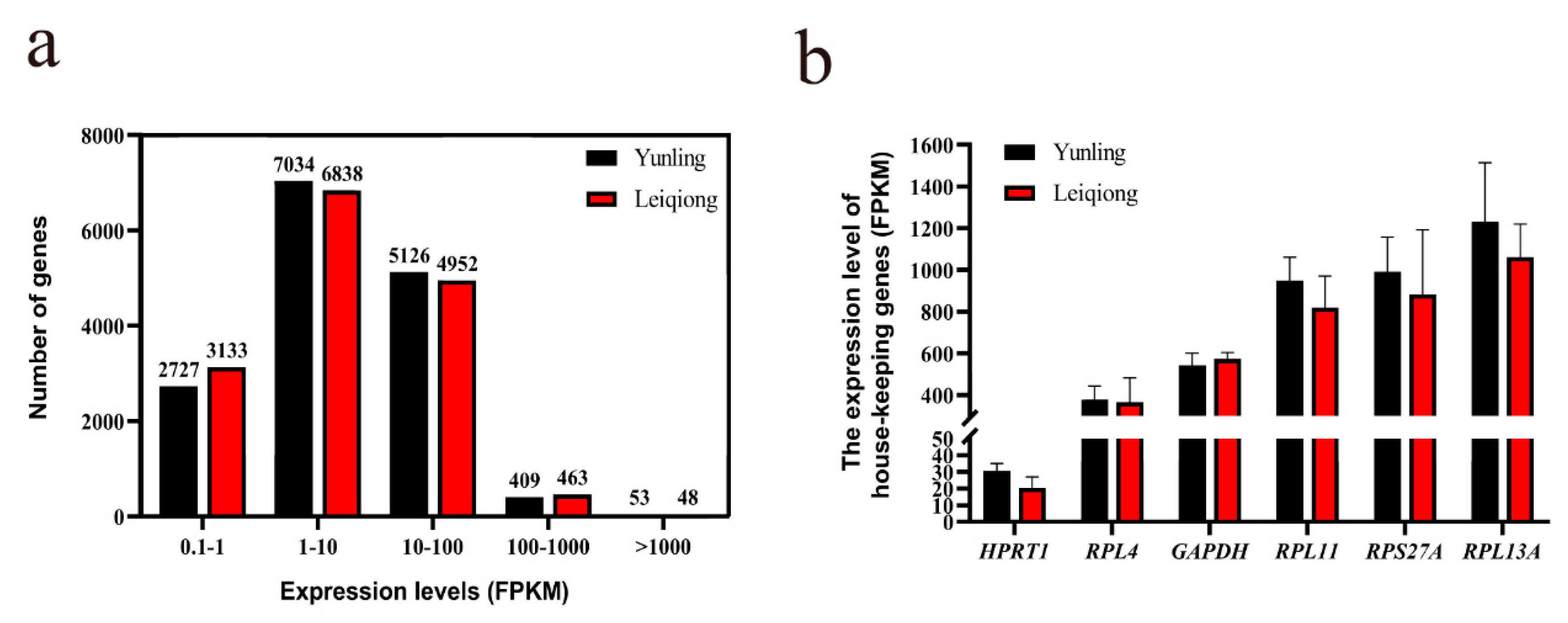
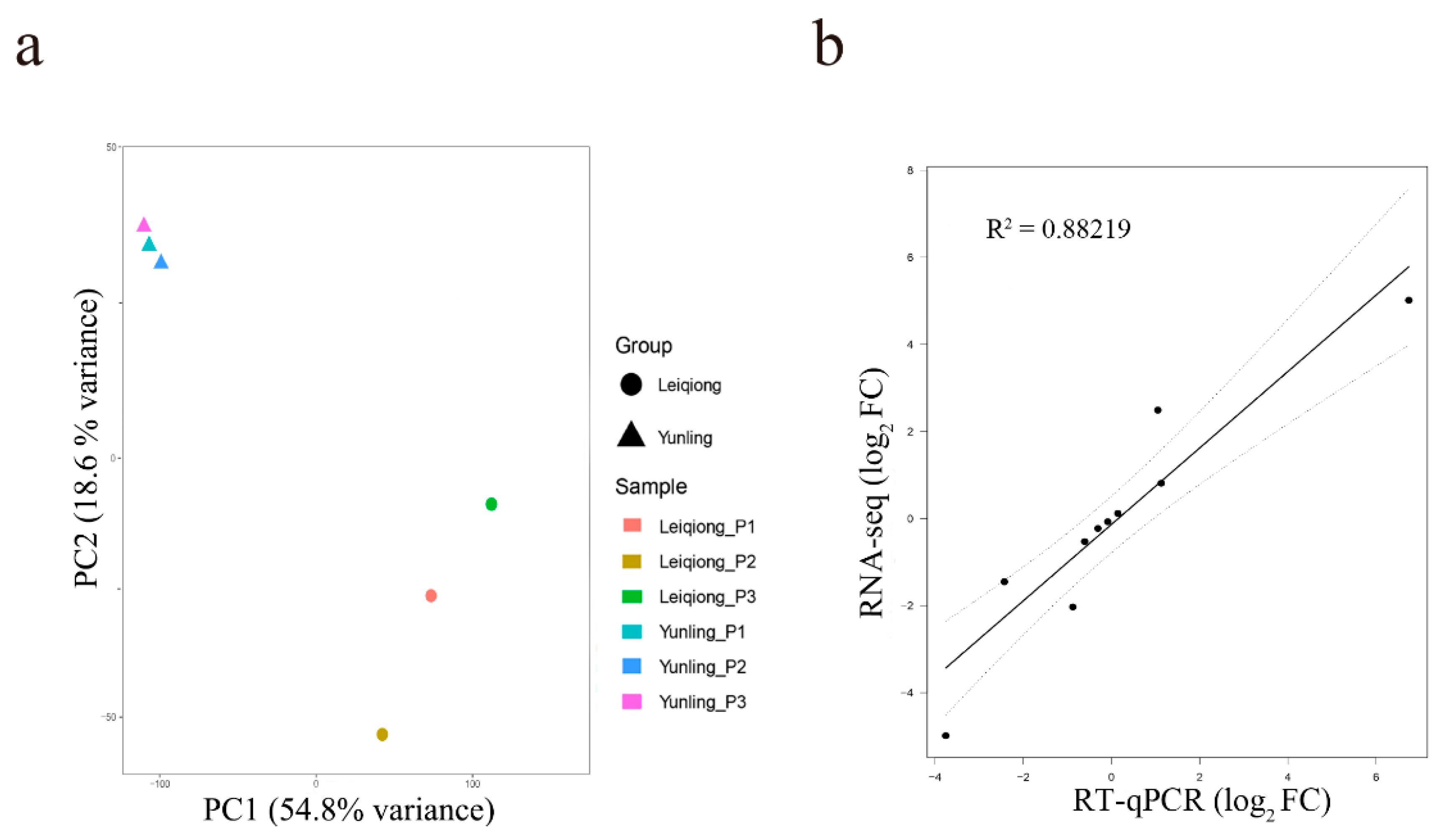


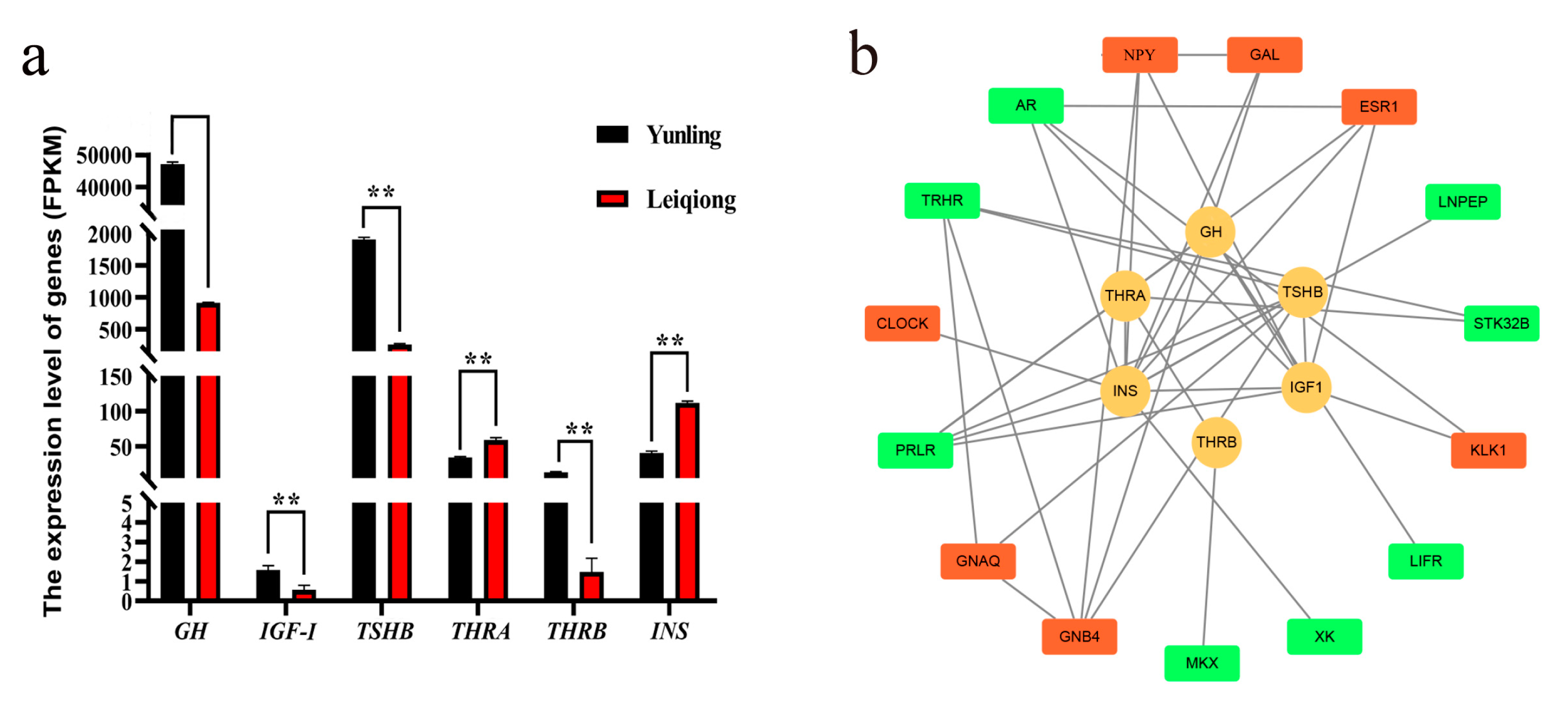
| Sample | Raw Reads | Clean Reads | Q30 | Total Mapped | Multiple Mapped | Uniquely Mapped |
|---|---|---|---|---|---|---|
| Yunling_P1 | 57,697,726 | 53,001,188 | 90.00% | 89.74% | 3.86% | 85.89% |
| Yunling_P2 | 58,487,994 | 53,293,322 | 89.72% | 89.42% | 4.81% | 84.62% |
| Yunling_P3 | 60,623,110 | 55,260,350 | 89.82% | 89.43% | 4.20% | 85.23% |
| Leiqiong_P1 | 65,019,640 | 59,273,088 | 84.65% | 66.21% | 2.25% | 63.96% |
| Leiqiong_P2 | 63,584,756 | 60,275,154 | 93.51% | 79.65% | 4.09% | 75.56% |
| Leiqiong_P3 | 60,204,686 | 56,734,662 | 92.89% | 78.26% | 3.53% | 74.73% |
| Pathway | Description | Gene Name | p-Value |
|---|---|---|---|
| bta04724 | Glutamatergic synapse | SLC38A1 → GNB4 → GRIN2A → SLC38A3 → GNAQ | 0.0011 |
| bta04961 | Endocrine and other factor-regulated calcium reabsorption | ESR1 → GNAQ → KLK1 | 0.0050 |
| bta04713 | Circadian entrainment | GNB4 → GRIN2A → GNAQ → GUCY1A2 | 0.0051 |
| bta04919 | Thyroid hormone signaling pathway | ESR1 → THRB → ITGAV → MED12L | 0.0091 |
| bta04080 | Neuroactive ligand–receptor interaction | NPY → GRIN2A → GAL → THRB → TRHR | 0.0127 |
| bta04614 | Renin-angiotensin system | KLK1 → LNPEP | 0.0140 |
| bta04728 | Dopaminergic synapse | GNB4 → GRIN2A → GNAQ → CLOCK | 0.0140 |
| bta00512 | Mucin type O-glycan biosynthesis | GCNT4 → GALNT5 | 0.0196 |
| bta04540 | Gap junction | MAP3K2 → GNAQ → GUCY1A2 | 0.0247 |
| bta04727 | GABAergic synapse | SLC38A1 → GNB4 → SLC38A3 | 0.0254 |
| bta00250 | Alanine, aspartate, and glutamate metabolism | AGXT2 → GFPT1 | 0.0273 |
| bta05017 | Spinocerebellar ataxia | GRIN2A → GNAQ → KCND3 | 0.0292 |
| bta00564 | Glycerophospholipid metabolism | LPGAT1 → ETNK1 → DGKH | 0.0358 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, X.; Arbab, A.A.I.; Zhang, Z.; Fan, Y.; Han, Z.; Gao, Q.; Sun, Y.; Yang, Z. Comparative Transcriptomic Analysis of the Pituitary Gland between Cattle Breeds Differing in Growth: Yunling Cattle and Leiqiong Cattle. Animals 2020, 10, 1271. https://doi.org/10.3390/ani10081271
Lu X, Arbab AAI, Zhang Z, Fan Y, Han Z, Gao Q, Sun Y, Yang Z. Comparative Transcriptomic Analysis of the Pituitary Gland between Cattle Breeds Differing in Growth: Yunling Cattle and Leiqiong Cattle. Animals. 2020; 10(8):1271. https://doi.org/10.3390/ani10081271
Chicago/Turabian StyleLu, Xubin, Abdelaziz Adam Idriss Arbab, Zhipeng Zhang, Yongliang Fan, Ziyin Han, Qisong Gao, Yujia Sun, and Zhangping Yang. 2020. "Comparative Transcriptomic Analysis of the Pituitary Gland between Cattle Breeds Differing in Growth: Yunling Cattle and Leiqiong Cattle" Animals 10, no. 8: 1271. https://doi.org/10.3390/ani10081271
APA StyleLu, X., Arbab, A. A. I., Zhang, Z., Fan, Y., Han, Z., Gao, Q., Sun, Y., & Yang, Z. (2020). Comparative Transcriptomic Analysis of the Pituitary Gland between Cattle Breeds Differing in Growth: Yunling Cattle and Leiqiong Cattle. Animals, 10(8), 1271. https://doi.org/10.3390/ani10081271





