Evaluating the Impact of Wildlife Shelter Management on the Genetic Diversity of Erinaceus europaeus and E. roumanicus in Their Contact Zone
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Sample Collection and DNA Isolation
2.2. Molecular Marker Enrichment and Amplification
2.3. Illumina Sequencing and Sequence Data Analysis
2.4. Population Genetic Analysis
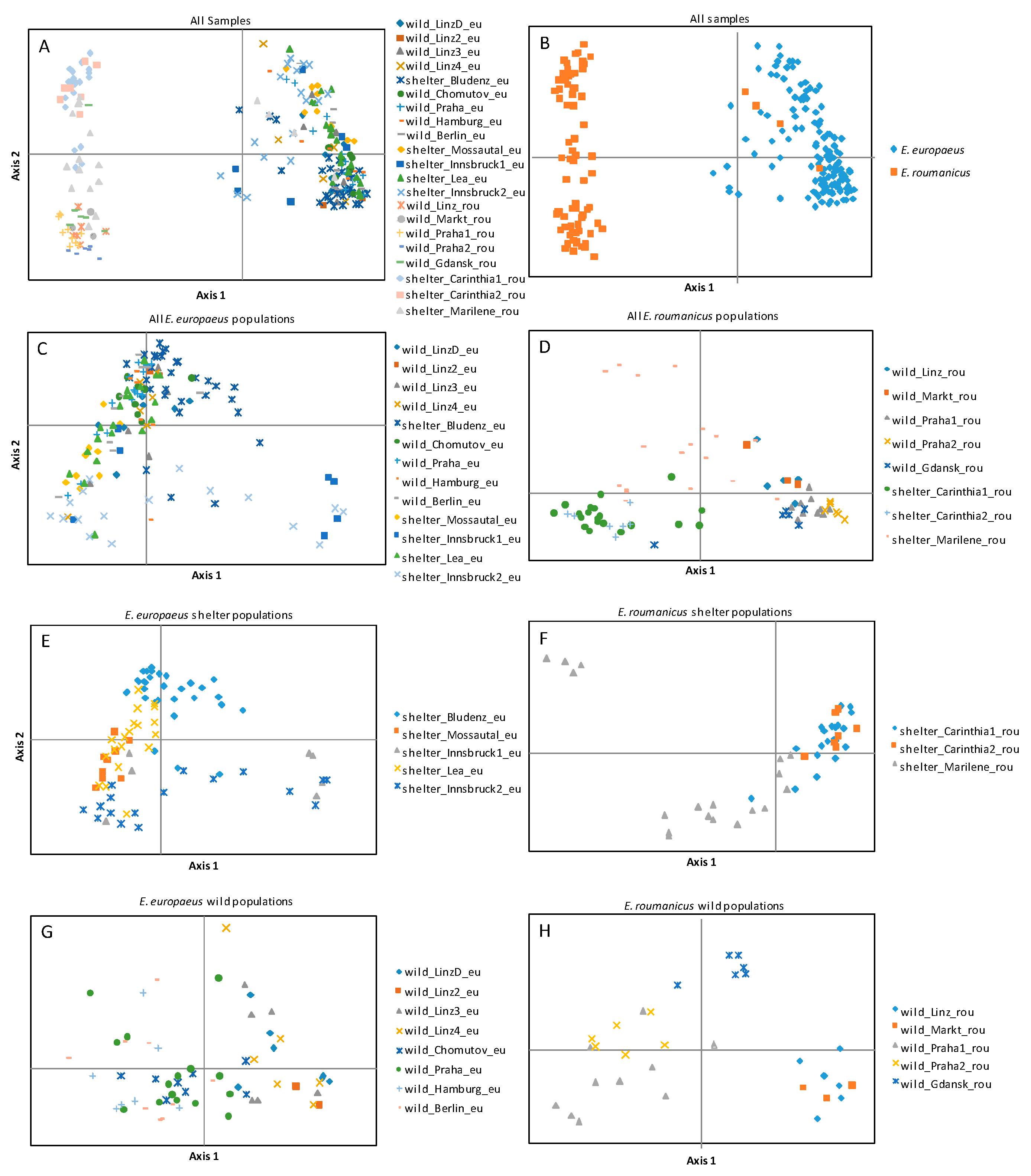
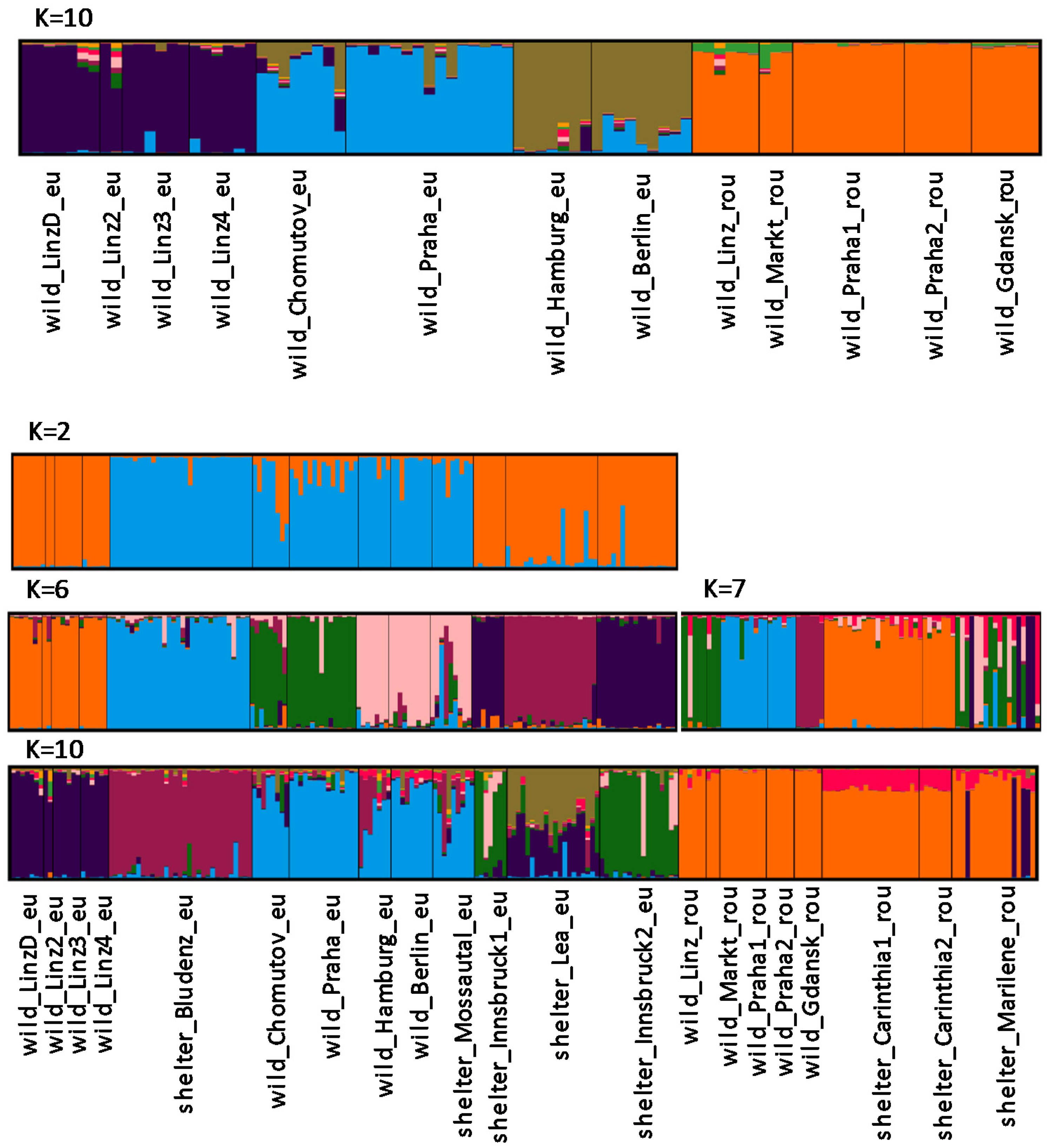
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Statement
References
- Amori, G. Erinaceus Europaeus. The IUCN Red List of Threatened Species 2016: E.T29650A2791303. In IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2016. [Google Scholar] [CrossRef]
- Amori, G.; Hutterer, R.; Kryštufek, B.; Yigit, N.; Mitsain, G.; Palomo, L.J. Erinaceus Roumanicus. The IUCN Red List of Threatened Species 2016: E.T136344A115206348. In IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2016. [Google Scholar] [CrossRef]
- Holsbeek, L.; Rodts, J.; Muyldermans, S. Hedgehog and other animal traffic victims in Belgium: Results of a countrywide survey. Lutra 1999, 42, 111–119. [Google Scholar]
- Huijser, M.P.; Bergers, P.J.M. The effect of roads and traffic on hedgehog (Erinaceus europaeus) populations. Biol. Conserv. 2000, 95, 111–116. [Google Scholar] [CrossRef]
- Hof, A.R.; Bright, P.W. Quantifying the long-term decline of the West European hedgehog in England by subsampling citizen-science datasets. Eur. J. Wildl. Res. 2016, 62, 407–413. [Google Scholar] [CrossRef]
- Hof, A.R. A Study of the Current Status of the Hedgehog (Erinaceus europaeus) and its Decline in Great Britain Since 1960. Ph.D. Thesis, Royal Holloway, University of London, Egham, UK, 2009. [Google Scholar]
- Becher, S.A.; Griffiths, R. Genetic differentiation among local populations of the European hedgehog (Erinaceus europaeus) in mosaic habitats. Mol. Ecol. 1998, 7, 1599–1604. [Google Scholar] [CrossRef] [PubMed]
- Černá Bolfíková, B.; Eliášová, K.; Loudová, M.; Kryštufek, B.; Lymberakis, P.; Sándor, A.D.; Hulva, P. Glacial allopatry vs. postglacial parapatry and peripatry: The case of hedgehogs. PeerJ 2017, 5, e3163. [Google Scholar] [CrossRef] [PubMed]
- Bolfíková, B.; Hulva, P. Microevolution of sympatry: Landscape genetics of hedgehogs Erinaceus europaeus and E. roumanicus in Central Europe. Heredity 2012, 108, 248–255. [Google Scholar] [CrossRef] [Green Version]
- Curto, M.; Winter, S.; Seiter, A.; Schmid, L.; Scheicher, K.; Barthel, L.M.F.; Plass, J.; Meimberg, H. Application of a SSR-GBS marker system on investigation of European Hedgehog species and their hybrid zone dynamics. Ecol. Evol. 2019, 9, 2814–2832. [Google Scholar] [CrossRef] [Green Version]
- Morris, P. Hedgehog; New Naturalist; Harper Collins: London, UK, 2018; Volume 137, ISBN 978-0-00-823573-4. [Google Scholar]
- Rondinini, C.; Doncaster, C.P. Roads as barriers to movement for hedgehogs. Funct. Ecol. 2002, 16, 504–509. [Google Scholar] [CrossRef]
- Orlowski, G.; Nowak, L. Road mortality of hedgehogs Erinaceus spp. in farmland in Lower Silesia (south-western Poland). Pol. J. Ecol. 2004, 52, 377–382. [Google Scholar]
- Braaker, S.; Kormann, U.; Bontadina, F.; Obrist, M.K. Prediction of genetic connectivity in urban ecosystems by combining detailed movement data, genetic data and multi-path modelling. Landsc. Urban Plan. 2017, 160, 107–114. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000; ISBN 978-0-674-66638-2. [Google Scholar]
- Avise, J.C. Phylogeography: Retrospect and prospect. J. Biogeogr. 2009, 36, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Molony, S.E.; Dowding, C.V.; Baker, P.J.; Cuthill, I.C.; Harris, S. The effect of translocation and temporary captivity on wildlife rehabilitation success: An experimental study using European hedgehogs (Erinaceus europaeus). Biol. Conserv. 2006, 130, 530–537. [Google Scholar] [CrossRef]
- Fuhrmann, M. Untersuchungen zur Wildtierrehabilitation und Wiederauswilderung von Erinaceus europaeus (Braunbrustigel) und Erinaceus roumanicus (Nördlicher Weißbrustigel) in Österreich. Master’s Thesis, University of Natural Resources and Life Sciences Vienna, Vienna, Austria, 2018. [Google Scholar]
- Weeks, A.R.; Sgro, C.M.; Young, A.G.; Frankham, R.; Mitchell, N.J.; Miller, K.A.; Byrne, M.; Coates, D.J.; Eldridge, M.D.B.; Sunnucks, P.; et al. Assessing the benefits and risks of translocations in changing environments: A genetic perspective. Evol. Appl. 2011, 4, 709–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allendorf, F.W.; Luikart, G.H.; Aitken, S.N. Conservation and the Genetics of Populations, 2nd ed.; Wiley–Blackwell: Hoboken, NJ, USA, 2013. [Google Scholar]
- Laikre, L.; Schwartz, M.K.; Waples, R.S.; Ryman, N. Compromising genetic diversity in the wild: Unmonitored large-scale release of plants and animals. Trends Ecol. Evol. 2010, 25, 520–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EUR-Lex: Access to European Union Law. Document L:1992:206:TOC. Available online: https://eur-lex.europa.eu/legal-content/DE/TXT/?uri=OJ:L:1992:206:TOC (accessed on 29 July 2020).
- Rechtsinformationssystem des Bundes RIS–Landesrecht. Available online: https://www.ris.bka.gv.at/Land/ (accessed on 31 July 2020).
- Edmands, S. Between a rock and a hard place: Evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 2007, 16, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.; Early, G.; Touhey, K.; Barco, S.; Gulland, F.; Wells, R. Rehabilitation and Release of Marine Mammals in the United States: Risks and Benefits. Mar. Mamm. Sci. 2007, 23, 731–750. [Google Scholar] [CrossRef]
- Seddon, J.M.; Santucci, F.; Reeve, N.J.; Hewitt, G.M. DNA footprints of European hedgehogs, Erinaceus europaeus and E. concolor: Pleistocene refugia, postglacial expansion and colonization routes. Mol. Ecol. 2001, 10, 2187–2198. [Google Scholar] [CrossRef] [Green Version]
- Suchentrunk, F.; Haiden, A.; Hartl, G.B. On biochemical genetic variability and divergence of the two hedgehog species Erinaceus europaeus and E. concolor in central Europe. Z. Säugetierkd. 1998, 63, 257–265. [Google Scholar]
- Hewitt, G.M. Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 1999, 68, 87–112. [Google Scholar] [CrossRef]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Santucci, F.; Emerson, B.C.; Hewitt, G.M. Mitochondrial DNA phylogeography of European hedgehogs. Mol. Ecol. 1998, 7, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.-G.; Cosson, J.-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Filippucci, M.G.; Simson, S. Allozyme Variation and Divergence in Erinaceidae (Mammalia, Insectivora). Isr. J. Zool. 1996, 42, 335–345. [Google Scholar] [CrossRef]
- Berggren, K.T.; Ellegren, H.; Hewitt, G.M.; Seddon, J.M. Understanding the phylogeographic patterns of European hedgehogs, Erinaceus concolor and E. europaeus using the MHC. Heredity 2005, 95, 84–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommer, R.S. When east met west: The sub-fossil footprints of the west European hedgehog and the northern white-breasted hedgehog during the Late Quaternary in Europe. J. Zool. 2007, 273, 82–89. [Google Scholar] [CrossRef]
- Tibihika, P.D.; Curto, M.; Dornstauder-Schrammel, E.; Winter, S.; Alemayehu, E.; Waidbacher, H.; Meimberg, H. Application of microsatellite genotyping by sequencing (SSR-GBS) to measure genetic diversity of the East African Oreochromis niloticus. Conserv. Genet. 2019, 20, 357–372. [Google Scholar] [CrossRef] [Green Version]
- Mcurto/SSR-GBS-Pipeline. Available online: https://github.com/mcurto/SSR-GBS-pipeline (accessed on 26 July 2020).
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar]
- Earl, D.A.; vonHoldt, B.M. Structure harvester: A website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Structure Harvester. Available online: http://taylor0.biology.ucla.edu/structureHarvester/ (accessed on 26 July 2020).
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [Green Version]
- CLUMPAK. Cluster Markov Packager Across K. Available online: http://clumpak.tau.ac.il/ (accessed on 26 July 2020).
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Europakarte—Die Karte von Europa. Available online: https://www.europakarte.org/ (accessed on 30 July 2020).
- Morris, P.A.; Meakin, K.; Sharafi, S. The Behaviour and Survival of Rehabilitated Hedgehogs (Erinaceus europaeus). Anim. Welf. 1993, 2, 53–66. [Google Scholar]
- Morris, P.A.; Warwick, H. A Study of Rehabilitated Juvenile Hedgehogs After Release into the Wild. Anim. Welf. 1994, 3, 163–177. [Google Scholar]
- Reeve, N.J. The Survival and Welfare of Hedgehogs (Erinaceus europaeus) After Release Back into the Wild. Anim. Welf. 1998, 7, 189–202. [Google Scholar]
- Bogdanov, A.S.; Bannikova, A.A.; Pirusskii, Y.M.; Formozov, N.A. The first genetic evidence of hybridization between West European and Northern White-breasted hedgehogs (Erinaceus europaeus and E. roumanicus) in Moscow Region. Biol. Bull. Russ. Acad. Sci. 2009, 36, 647–651. [Google Scholar] [CrossRef]
- Bolfíková, B.; Konečný, A.; Pfäffle, M.; Skuballa, J.; Hulva, P. Population biology of establishment in New Zealand hedgehogs inferred from genetic and historical data: Conflict or compromise? Mol. Ecol. 2013, 22, 3709–3720. [Google Scholar] [CrossRef]


| Population | # Individuals | Species | Geographical Origin |
|---|---|---|---|
| wild_LinzD_eu | 7 | E. europaeus | Linz/AUT (wild) |
| wild_Linz2_eu | 2 | E. europaeus | Linz/AUT (wild) |
| wild_Linz3_eu | 6 | E. europaeus | Linz/AUT (wild) |
| wild_Linz4_eu | 6 | E. europaeus | Linz/AUT (wild) |
| shelter_Bludenz_eu | 31 | E. europaeus | Bludenz/AUT (shelter) |
| wild_Chomutov_eu | 8 | E. europaeus | Chomutov/CZE (wild) |
| wild_Praha_eu | 15 | E. europaeus | Prague/CZE (wild) |
| wild_Hamburg_eu | 7 | E. europaeus | Hamburg/GER (wild) |
| wild_Berlin_eu | 9 | E. europaeus | Berlin/GER (wild) |
| shelter_Mossautal_eu | 9 | E. europaeus | Mossautal/GER (shelter) |
| shelter_Innsbruck1_eu | 7 | E. europaeus | Innsbruck/AUT (shelter) |
| shelter_Lea_eu | 20 | E. europaeus | Bavaria/GER (shelter) |
| shelter_Innsbruck2_eu | 17 | E. europaeus | Innsbruck/AUT (shelter) |
| wild_Linz_rou | 6 | E. roumanicus | Linz/AUT (wild) |
| wild_Markt_rou | 3 | E. roumanicus | Linz/AUT (wild |
| wild_Praha1_rou | 10 | E. roumanicus | Prague/CZE (wild) |
| wild_Praha2_rou | 6 | E. roumanicus | Prague/CZE (wild) |
| wild_Gdansk_rou | 6 | E. roumanicus | Gdansk/POL (wild) |
| shelter_Carinthia1_rou | 21 | E. roumanicus | Carinthia/AUT (shelter) |
| shelter_Carinthia2_rou | 7 | E. roumanicus | Carinthia/AUT (shelter) |
| shelter_Marilene_rou | 18 | E. roumanicus | Graz/AUT (shelter) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ploi, K.; Curto, M.; Bolfíková, B.Č.; Loudová, M.; Hulva, P.; Seiter, A.; Fuhrmann, M.; Winter, S.; Meimberg, H. Evaluating the Impact of Wildlife Shelter Management on the Genetic Diversity of Erinaceus europaeus and E. roumanicus in Their Contact Zone. Animals 2020, 10, 1452. https://doi.org/10.3390/ani10091452
Ploi K, Curto M, Bolfíková BČ, Loudová M, Hulva P, Seiter A, Fuhrmann M, Winter S, Meimberg H. Evaluating the Impact of Wildlife Shelter Management on the Genetic Diversity of Erinaceus europaeus and E. roumanicus in Their Contact Zone. Animals. 2020; 10(9):1452. https://doi.org/10.3390/ani10091452
Chicago/Turabian StylePloi, Kerstin, Manuel Curto, Barbora Černá Bolfíková, Miroslava Loudová, Pavel Hulva, Anna Seiter, Marilene Fuhrmann, Silvia Winter, and Harald Meimberg. 2020. "Evaluating the Impact of Wildlife Shelter Management on the Genetic Diversity of Erinaceus europaeus and E. roumanicus in Their Contact Zone" Animals 10, no. 9: 1452. https://doi.org/10.3390/ani10091452





