CircRNA-1926 Promotes the Differentiation of Goat SHF Stem Cells into Hair Follicle Lineage by miR-148a/b-3p/CDK19 Axis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Skin Tissues of Goats
2.2. Sequence Analysis of CircRNA-1926
2.3. Cell Cultivation and Overexpression/siRNA Interference Analysis of CircRNA-1926
2.4. RNA Pull-Down Assay
2.5. Extraction of Total RNA and Real-Time PCR Reactions
2.6. Methylation Detection of CDK19 Gene Promoter in SHF Stem Cells
2.7. Dual-Luciferase Reporter Assays
2.8. Statistical Analysis
3. Results and Discussion
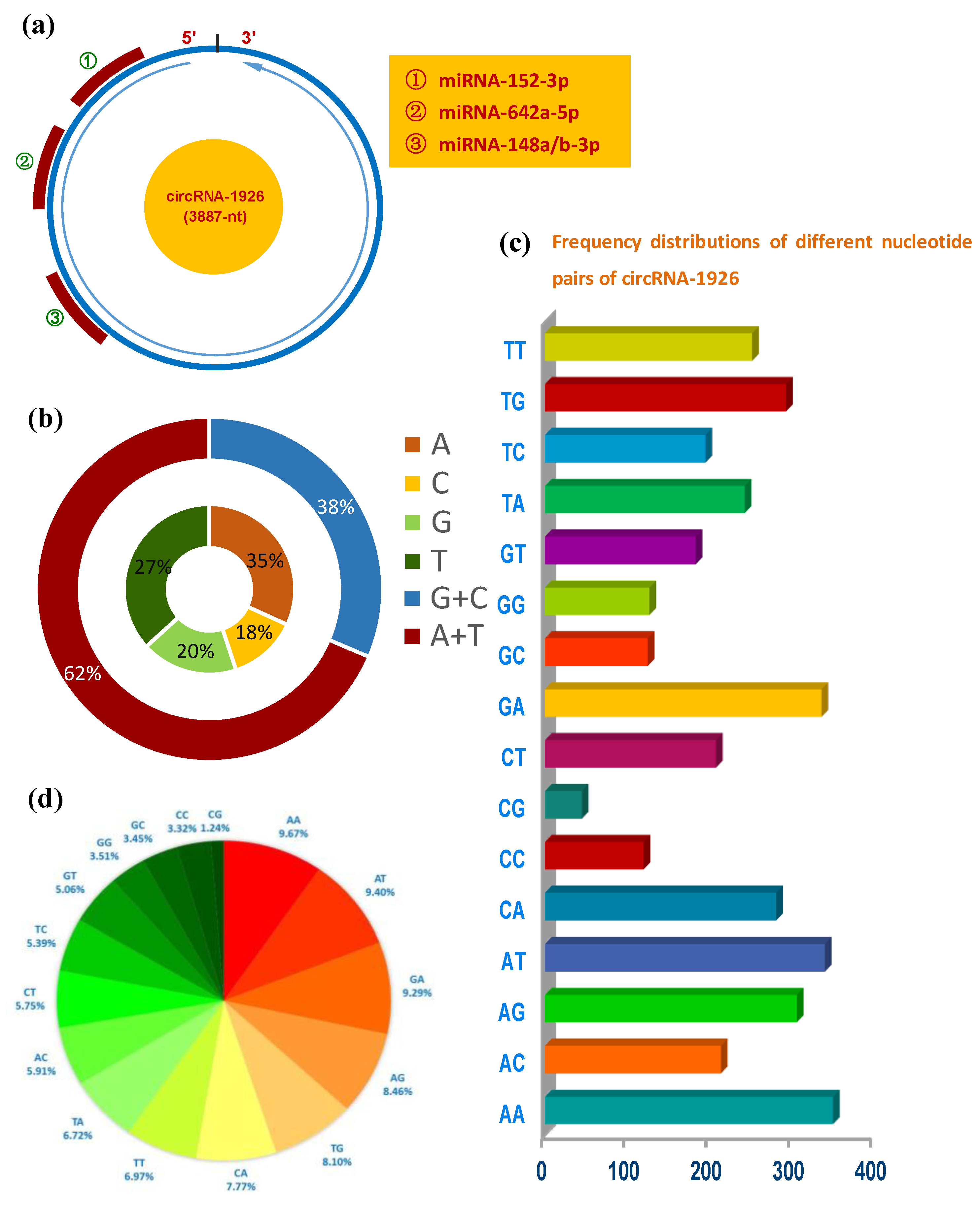
3.1. Sequence Analysis of CircRNA-1926 in Cashmere Goat SHF
3.2. Expression Analysis of CircRNA-1926 in Bulge and Its Role in Regulating the Differentiation of SHF Stem Cells into Hair Follicle Lineages
3.3. CircRNA-1926 Directly Combines with miR-148a/b-3p and May Regulate Their Expression in SHF Stem Cells
3.4. CircRNA-1926 Promotes CDK19 Expression without Modulating the Methylation Level of Its Promoter Region
3.5. CircRNA-1926 Positively Regulates the Expression of CDK19 through miR-148a/b-3p
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, G.; Kang, D.; Ma, S.; Wang, X.; Gao, Y.; Yang, Y.; Wang, X.; Chen, Y. Integrative analysis reveals ncRNA-mediated molecular regulatory network driving secondary hair follicle regression in cashmere goats. BMC Genom. 2018, 19, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, Q.; Yin, R.H.; Zhao, S.J.; Wang, Z.Y.; Zhu, Y.B.; Wang, W.; Zheng, Y.Y.; Yin, X.B.; Guo, D.; Wang, S.Q.; et al. Identification and molecular analysis of a lncRNA-HOTAIR transcript from secondary hair follicle of cashmere goat reveal integrated regulatory network with the expression regulated potentially by its promoter methylation. Gene 2019, 688, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, Y.; Zhang, J.; Chang, Z.; Li, J.; Yan, Z.; Zhang, W. Hoxc13/β-catenin correlation with hair follicle activity in Cashmere goat. J. Integr. Agric. 2012, 11, 1159–1166. [Google Scholar] [CrossRef]
- Wang, X.; Yin, J. Indirect location of hair follicle stem cell nests in Inner Mongolia white cashmere goat. J. Agric. Biotechnol. 2014, 22, 326–332. [Google Scholar]
- Morgan, B.A. The dermal papilla: An instructive niche for epithelial stem and progenitor cells in development and regeneration of the hair follicle. Cold Spring Harb. Perspect. Med. 2014, 4, a015180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, Y.; Bai, J.; Wu, J.; Li, Q.; Mo, Y.; Fang, R.; Lai, W. LncRNA PlncRNA-1 regulates proliferation and differentiation of hair follicle stem cells through TGF-β1-mediated Wnt/β-catenin signal pathway. Mol. Med. Rep. 2018, 17, 1191–1197. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Shi, C.; Wang, Y.; Yang, J.; Yang, T. Regulatory effect of β-catenin on proliferation of hair follicle stem cells involves pi3k/akt pathway. J. Appl. Biomed. 2013, 11, 131–141. [Google Scholar] [CrossRef]
- Folgueras, A.R.; Guo, X.; Pasolli, H.A.; Stokes, N.; Polak, L.; Zheng, D.; Fuchs, E. Architectural niche organization by LHX2 is linked to hair follicle stem cell function. Cell Stem Cell 2013, 13, 314–327. [Google Scholar] [CrossRef] [Green Version]
- Amelio, I.; Lena, A.M.; Bonanno, E.; Melino, G.; Candi, E. miR-24 affects hair follicle morphogenesis targeting Tcf-3. Cell Death Dis. 2013, 4, e922. [Google Scholar] [CrossRef] [Green Version]
- Merrill, B.J.; Gat, U.; DasGupta, R.; Fuchs, E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001, 15, 1688–1705. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xing, Y.; Guo, H.; Ma, X.; Li, Y. Immunohistochemical study of hair follicle stem cells in regenerated hair follicles induced by Wnt10b. Int. J. Med. Sci. 2016, 13, 765–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Q.; Yu, W.; Fang, Y.; Yao, M.; Yang, P. Beta-catenin can induce hair follicle stem cell differentiation into transit-amplifying cells through c-myc activation. Tissue Cell 2017, 49, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Yuan, C.; He, X.; Kang, D.; Wang, X.; Chen, Y. Effect of mir-125b on dermal papilla cells of goat secondary hair follicle. Electron. J. Biotechnol. 2017, 25, 64–69. [Google Scholar] [CrossRef]
- Zhu, Y.B.; Wang, Z.Y.; Yin, R.H.; Jiao, Q.; Zhao, S.J.; Cong, Y.Y.; Xue, H.L.; Guo, D.; Wang, S.Q.; Zhu, Y.X.; et al. A lncRNA-H19 transcript from secondary hair follicle of Liaoning cashmere goat: Identification, regulatory network and expression regulated potentially by its promoter methylation. Gene 2018, 641, 78–85. [Google Scholar] [CrossRef]
- Ohyama, M.; Kobayashi, T. Isolation and characterization of stem cell-enriched human and canine hair follicle keratinocytes. Methods Mol. Biol. 2012, 879, 389–401. [Google Scholar] [PubMed]
- Yin, R.; Wang, Y.; Wang, Z.; Zhu, Y.; Cong, Y.; Wang, W.; Deng, L.; Liu, H.; Guo, D.; Bai, W. Discovery and molecular analysis of conserved circRNAs from cashmere goat reveal their integrated regulatory network and potential roles in secondary hair follicle. Electron. J. Biotechn. 2019, 41, 37–47. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Gao, Y.; Ding, Q.; Liu, J.; Li, Y.; Jin, M.; Xu, H.; Ma, S.; Wang, X.; Zeng, W.; et al. Exosomal micro rnas derived from dermal papilla cells mediate hair follicle stem cell proliferation and differentiation. Int. J. Biol. Sci. 2019, 15, 1368–1382. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Chao, Y.; Zhou, G.; Chen, Y. Fibroblast growth factor 5-short (FGF5s) inhibits the activity of FGF5 in primary and secondary hair follicle dermal papilla cells of cashmere goats. Gene 2016, 575, 393–398. [Google Scholar] [CrossRef]
- He., J.; Huang, Z.; He, M.; Liao, J.; Zhang, Q.; Wang, S.; Xie, L.; Ouyang, L.; Koeffler, H.P.; Yin, D.; et al. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol. Cancer 2020, 19, 17. [Google Scholar] [CrossRef]
- Kumaki, Y.; Oda, M.; Okano, M. QUMA: Quantification tool for methylation analysis. Nucleic Acids Res. 2008, 36, W170–W175. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Li, L.; Xie, F.; Guo, S.; Liu, F.; Dong, N.; Wang, Y. LncRNA TUG1 sponges miR-204–5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc. Res. 2018, 114, 168–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, W.L.; Yin, R.H.; Yin, R.L.; Jiang, W.Q.; Wang, J.J.; Wang, Z.Y.; Zhu, Y.B.; Zhao, Z.H.; Yang, R.J.; Luo, G.B.; et al. Selection and validation of suitable reference genes in skin tissue of Liaoning Cashmere goat during hair follicle cycle. Livest. Sci. 2014, 161, 28–35. [Google Scholar] [CrossRef]
- Bai, W.L.; Dang, Y.L.; Yin, R.H.; Yin, R.L.; Luo, G.B. Combination of let-7d-5p, mir-26a-5p, and mir-15a-5p is suitable normalizer for studying microrna expression in skin tissue of liaoning cashmere goat during hair follicle cycle. Czech J. Anim. Sci. 2016, 61, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Haerty, W.; Ponting, C.P. Unexpected selection to retain high GC content and splicing enhancers within exons of multiexonic lncRNA loci. RNA 2015, 21, 333–346. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Kennedy, S.D.; Turner, D.H. A CA(+) pair adjacent to a sheared GA or AA pair stabilizes size-symmetric RNA internal loops. Biochemistry 2009, 48, 5738–5752. [Google Scholar] [CrossRef]
- Lagnado, C.A.; Brown, C.Y.; Goodall, G.J. AUUUA Is Not Sufficient to Promote poly(A) Shortening and Degradation of an mRNA: The Functional Sequence Within AU-rich Elements May Be UUAUUUA(U/A)(U/A). Mol. Cell. Biol. 1994, 14, 7984–7995. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [Green Version]
- Ducongé, F.; Di Primo, C.; Toulme, J.J. Is a closing “GA pair” a rule for stable loop-loop RNA complexes? J. Biol. Chem. 2000, 275, 21287–21294. [Google Scholar] [CrossRef] [Green Version]
- Paus, K.; Muller-Rover, S. Comprehensive guide for the recognition classification of distinct stages of hair follicle morphogenesis. J. Investig. Dermatol. 1999, 113, 523–532. [Google Scholar]
- Wang, S.; Ge, W.; Luo, Z.; Guo, Y.; Jiao, B.; Qu, L.; Zhang, Z.; Wang, X. Integrated analysis of coding genes and non-coding RNAs during hair follicle cycle of cashmere goat (Capra hircus). BMC Genom. 2017, 18, 767. [Google Scholar] [CrossRef] [Green Version]
- Misago, N.; Narisawa, Y. Tricholemmal carcinoma in continuity with trichoblastoma within nevus sebaceous. Am. J. Dermatopathol. 2002, 24, 149–155. [Google Scholar] [CrossRef]
- Arumugam, A.; Weng, Z.; Chaudhary, S.C.; Afaq, F.; Elmets, C.A.; Athar, M. Keratin-6 driven ODC expression to hair follicle keratinocytes enhances stemness and tumorigenesis by negatively regulating Notch. Biochem. Biophys. Res. Commun. 2014, 451, 394–401. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Xie, G.; Zhu, L.; Chen, X.; Li, X.; Lu, H.; Xu, B.; Ramot, Y.; Paus, R.; Yue, Z. p53 is a direct transcriptional repressor of keratin 17: Lessons from a Rat model of radiation dermatitis. J. Investig. Dermatol. 2016, 136, 680–689. [Google Scholar] [CrossRef] [Green Version]
- Panteleyev, A.A.; Paus, R.; Wanner, R.; Nürnberg, W.; Eichmüller, S.; Thiel, R.; Zhang, J.; Henz, B.M.; Rosenbach, T. Keratin 17 gene expression during the murine hair cycle. J. Investig. Dermatol. 1997, 108, 324–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef]
- Han, K.; Wang, F.W.; Cao, C.H.; Ling, H.; Chen, J.W.; Chen, R.X.; Feng, Z.H.; Luo, J.; Jin, X.H.; Duan, J.L.; et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol. Cancer 2020, 19, 60. [Google Scholar] [CrossRef] [Green Version]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memcza, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Shan, G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription 2015, 6, 61–64. [Google Scholar] [CrossRef]
- Daugela, L.; Nüsgen, N.; Walier, M.; Oldenburg, J.; Schwaab, R.; El-Maarri, O. Measurements of DNA methylation at seven loci in various tissues of CD1 mice. PLoS ONE 2012, 7, e44585. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.Y.; Huang, X.J.; Chim, C.S. DNA methylation of microRNA genes in multiple myeloma. Carcinogenesis 2012, 33, 1629–1638. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, C.; Wu, Z.; Chen, Y.; Shi, W. CircIBTK inhibits DNA demethylation and activation of AKT signaling pathway via miR-29b in peripheral blood mononuclear cells in systemic lupus erythematosus. Arthritis Res. Ther. 2018, 20, 118. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Yu, Y.; Huang, Z.; Kong, Y.; Hu, X.; Xiao, W.; Quan, J.; Fan, X. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019, 10, 900. [Google Scholar] [CrossRef]
- Liu, H.; Xue, L.; Song, C.; Liu, F.; Jiang, T.; Yang, X. Overexpression of circular RNA circ_001569 indicates poor prognosis in hepatocellular carcinoma and promotes cell growth and metastasis by sponging miR-411-5p and miR-432-5p. Biochem. Biophys. Res. Commun. 2018, 503, 2659–2665. [Google Scholar] [CrossRef]
- Li, G.; Huang, M.; Cai, Y.; Yang, Y.; Sun, X.; Ke, Y. Circ-U2AF1 promotes human glioma via derepressing neuro-oncological ventral antigen 2 by sponging hsa-miR-7-5p. J. Cell. Physiol. 2019, 234, 9144–9155. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, G.; Fan, L.; Zhang, G.; Xu, J.; Zhang, J. Circular RNA circ_0034642 elevates BATF3 expression and promotes cell proliferation and invasion through miR-1205 in glioma. Biochem. Biophys. Res. Commun. 2019, 508, 980–985. [Google Scholar] [CrossRef] [PubMed]
- Dale, T.; Clarke, P.A.; Esdar, C.; Waalboer, D.; Adeniji-Popoola, O.; Ortiz-Ruiz, M.J.; Mallinger, A.; Samant, R.S.; Czodrowski, P.; Musil, D.; et al. A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease. Nat. Chem. Biol. 2015, 11, 973–980. [Google Scholar] [CrossRef]
- Firestein, R.; Bass, A.J.; Kim, S.Y.; Dunn, I.F.; Silver, S.J.; Guney, I.; Freed, E.; Ligon, A.H.; Vena, N.; Ogino, S.; et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 2008, 455, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Lien, W.H.; Polak, L.; Lin, M.; Lay, K.; Zheng, D.; Fuchs, E. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat. Cell Biol. 2014, 16, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yu, J.; Shi, C.; Huang, Y.; Wang, Y.; Yang, T.; Yang, J. Lef1 contributes to the differentiation of bulge stem cells by nuclear translocation and cross-talk with the Notch signaling pathway. Int. J. Med. Sci. 2013, 10, 738–746. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.S.; Zhang, Y.; Xu, M.; Yang, Y.; Ito, M.; Peng, T.; Zheng, C.; Nagy, A.; Hadjantonakis, A.; Lang, R.A.; et al. Distinct Functions for Wnt/β-Catenin in Hair Follicle Stem Cell Proliferation and Survival and Interfollicular Epidermal Homeostasis. Cell Stem Cell 2013, 13, 720–733. [Google Scholar] [CrossRef] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, R.H.; Zhao, S.J.; Jiao, Q.; Wang, Z.Y.; Bai, M.; Fan, Y.X.; Zhu, Y.B.; Bai, W.L. CircRNA-1926 Promotes the Differentiation of Goat SHF Stem Cells into Hair Follicle Lineage by miR-148a/b-3p/CDK19 Axis. Animals 2020, 10, 1552. https://doi.org/10.3390/ani10091552
Yin RH, Zhao SJ, Jiao Q, Wang ZY, Bai M, Fan YX, Zhu YB, Bai WL. CircRNA-1926 Promotes the Differentiation of Goat SHF Stem Cells into Hair Follicle Lineage by miR-148a/b-3p/CDK19 Axis. Animals. 2020; 10(9):1552. https://doi.org/10.3390/ani10091552
Chicago/Turabian StyleYin, Rong H., Su J. Zhao, Qian Jiao, Ze Y. Wang, Man Bai, Yi X. Fan, Yu B. Zhu, and Wen L. Bai. 2020. "CircRNA-1926 Promotes the Differentiation of Goat SHF Stem Cells into Hair Follicle Lineage by miR-148a/b-3p/CDK19 Axis" Animals 10, no. 9: 1552. https://doi.org/10.3390/ani10091552



