Variation of Gut Microbiome in Free-Ranging Female Tibetan Macaques (Macaca thibetana) across Different Reproductive States
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subject and Sample Collection
2.2. DNA Extraction and Sequencing
2.3. Sequence Analysis
2.4. Data Analysis
2.5. Data Availability
3. Results
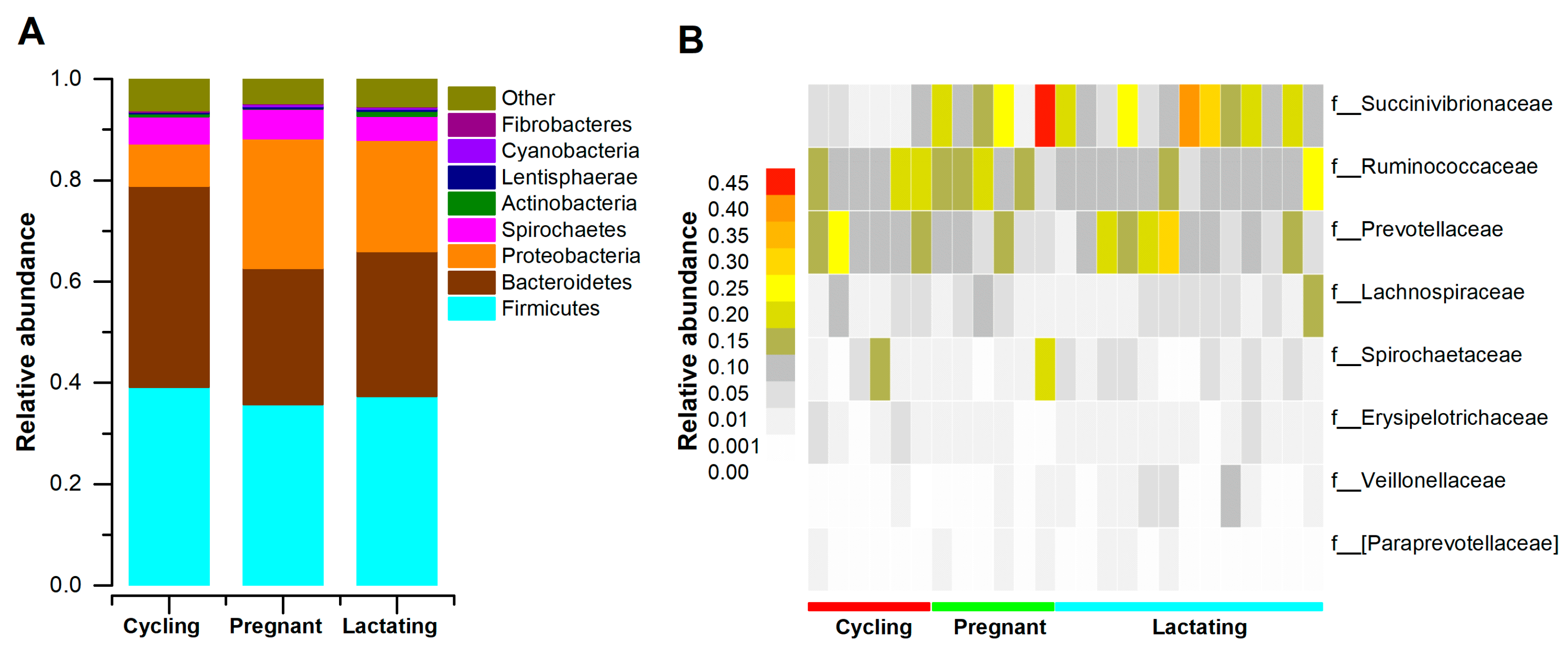
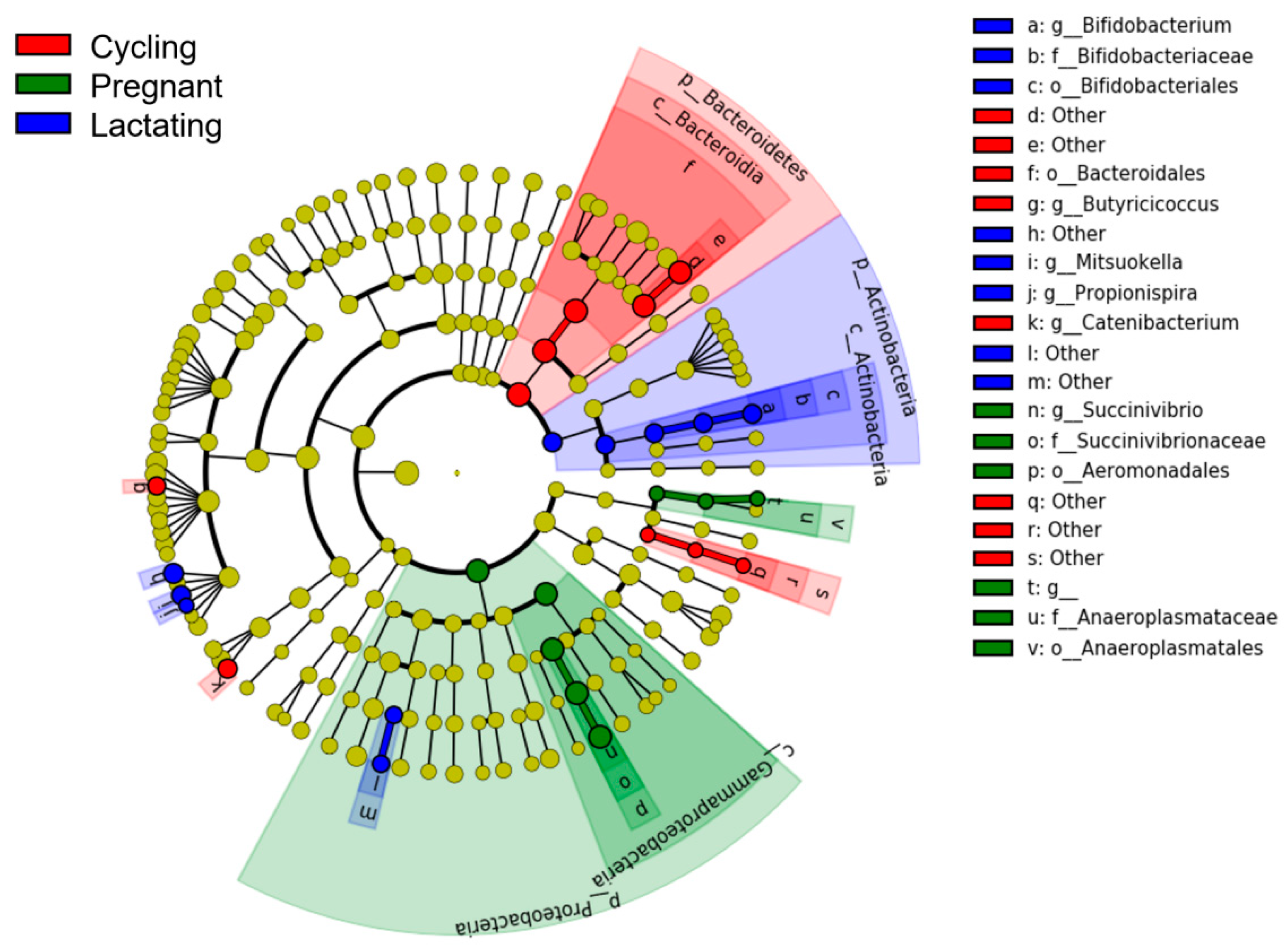
3.1. Microbial Community Profiles
3.2. Alpha and Beta Diversity
3.3. Predicted Metabolic Functions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gittleman, J.L.; Thompson, S.D. Energy Allocation in Mammalian Reproduction. Integr. Comp. Biol. 1988, 28, 863–875. [Google Scholar] [CrossRef]
- Oftedal, O.T.; Whiten, A.; Southgate, D.A.T.; Soest, P.V. The nutritional consequences of foraging in primates: the relationship of nutrient intakes to nutrient requirements. Philos. Trans. Biol. Sci. 1991, 334, 161–170. [Google Scholar]
- Wells, A.J.C.K. Energetics and the Evolution of the Genus HOMO. Annu. Rev. Anthropol. 2002, 31, 323–338. [Google Scholar]
- Miller, S.C.; Halloran, B.P.; Deluca, H.F.; Jee, W.S.S. Role of vitamin D in maternal skeletal changes during pregnancy and lactation: A histomorphometric study. Calcif. Tissue Int. 1982, 34, 245–252. [Google Scholar] [CrossRef]
- Thompson, M.E. Reproductive Ecology of Female Chimpanzees. Am. J. Primatol. 2013, 75, 222–237. [Google Scholar] [CrossRef]
- Mallott, E.K.; Amato, K.R. The microbial reproductive ecology of white-faced capuchins (Cebus capucinus). Am. J. Primatol. 2018, 80, e22896. [Google Scholar] [CrossRef]
- Key, C.; Ross, C. Sex differences in energy expenditure in non–human primates. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1999, 266, 2479–2485. [Google Scholar] [CrossRef] [Green Version]
- Rothman, J.M.; Raubenheimer, D.; Bryer, M.A.; Takahashi, M.; Gilbert, C.C. Nutritional contributions of insects to primate diets: implications for primate evolution. J. Hum. Evol. 2014, 71, 59–69. [Google Scholar] [CrossRef]
- Kurta, A.; Bell, G.P.; Nagy, K.A.; Kunz, T.H. Energetics of pregnancy and lactation in freeranging little brown bats (Myotis lucifugus). Physiol. Zool. 1989, 62, 804–818. [Google Scholar] [CrossRef]
- Dufour, D.L.; Sauther, M.L. Comparative and evolutionary dimensions of the energetics of human pregnancy and lactation. Am. J. Hum. Biol. 2002, 14, 584–602. [Google Scholar] [CrossRef]
- Bell, A.W.; Bauman, D.E. Adaptations of glucose metabolism during pregnancy and lactation. J. Mammary Gland. Biol. Neoplasia 1997, 2, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Bronson, F. Climate change and seasonal reproduction in mammals. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 3331–3340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooper, L.V.; Midtvedt, T.; Gordon, J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 2002, 22, 283–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [Green Version]
- Chevalier, C.; Stojanović, O.; Colin, D.J.; Suarez-Zamorano, N.; Tarallo, V.; Veyrat-Durebex, C.; Rigo, D.; Fabbiano, S.; Stevanović, A.; Hagemann, S. Gut microbiota orchestrates energy homeostasis during cold. Cell 2015, 163, 1360–1374. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Backhed, H.K.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Mallott, E.K.; Borries, C.; Koenig, A.; Amato, K.R.; Lu, A. Reproductive hormones mediate changes in the gut microbiome during pregnancy and lactation in Phayre’s leaf monkeys. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Liu, H.; Hou, C.; Li, N.; Zhang, X.; Zhang, G.; Yang, F.; Zeng, X.; Liu, Z.; Qiao, S. Microbial and metabolic alterations in gut microbiota of sows during pregnancy and lactation. FASEB J. 2019, 33, 4490–4501. [Google Scholar] [CrossRef]
- Julie, L.; Sébastien, M.; Hélène, S.; Clément, G.; Frédéric, D.; Martine, S.; Li, B.; José, W.; JérMe, B.; Nadia, E. Feeding sows resistant starch during gestation and lactation impacts their faecal microbiota and milk composition but shows limited effects on their progeny. PLoS ONE 2018, 13, e0199568. [Google Scholar]
- Jack, F. Ecogeographic segregation of macaque species. Primates 1982, 23, 574–579. [Google Scholar]
- Li, J.-H.; Kappeler, P.M. Social and Life History Strategies of Tibetan Macaques at Mt. Huangshan. In The Behavioral Ecology of the Tibetan Macaque; Springer: Berlin/Heidelberg, Germany, 2020; pp. 17–46. [Google Scholar]
- Sun, B.; Gu, Z.; Wang, X.; Huffman, M.A.; Garber, P.A.; Sheeran, L.K.; Zhang, D.; Zhu, Y.; Xia, D.P.; Li, J.H. Season, age, and sex affect the fecal mycobiota of free-ranging Tibetan macaques (Macaca thibetana). Am. J. Primatol. 2018, 80, e22880. [Google Scholar] [CrossRef] [PubMed]
- Thierry, B. The macaques: A double-layered social organization. In Primates in Perspective; Oxford University Press: Oxford, UK, 2007; pp. 224–239. [Google Scholar]
- Mori, H.; Maruyama, F.; Kato, H.; Toyoda, A.; Dozono, A.; Ohtsubo, Y.; Nagata, Y.; Fujiyama, A.; Tsuda, M.; Kurokawa, K. Design and Experimental Application of a Novel Non-Degenerate Universal Primer Set that Amplifies Prokaryotic 16S rRNA Genes with a Low Possibility to Amplify Eukaryotic rRNA Genes. DNA Res. 2014, 21, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.J.; Li, H.; Xie, P.; Li, Z.H.; Li, H.W.; Yin, Y.; Blachier, F.; Kong, X. Stages of pregnancy and weaning influence the gut microbiota diversity and function in sows. J. Appl. Microbiol. 2019, 127, 867–879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, I.; Azhar, E.I.; Abbas, A.T.; Kumosani, T.A.; Barbour, E.K.; Raoult, D.; Yasir, M. Metagenomic Analysis of Antibiotic-Induced Changes in Gut Microbiota in a Pregnant Rat Model. Front. Pharmacol. 2016, 7, 104. [Google Scholar] [CrossRef] [Green Version]
- Phillips, C.D.; Phelan, G.; Dowd, S.E.; Mcdonough, M.M.; Ferguson, A.W.; Hanson, J.D.; Siles, L.; Ordóez-Garza, N.; Francisco, M.S.; Baker, R.J. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol. Ecol. 2012, 21, 2617–2627. [Google Scholar] [CrossRef]
- Trevelline, B.K.; MacLeod, K.J.; Langkilde, T.; Kohl, K.D. Gestation alters the gut microbiota of an oviparous lizard. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef]
- Marlies, E.; Floor, H.; Clara, B.; Mark, B.; Bart, D.H.; Paul, D.V.; Marijke, F. Changes in intestinal gene expression and microbiota composition during late pregnancy are mouse strain dependent. Sci. Rep. 2018, 8, 10001. [Google Scholar]
- Krishna, A.; Bhatnagar, K.P. Hormones and Reproductive Cycles in Bats. In Hormones and Reproduction of Vertebrates; Elsevier Inc.: Amsterdam, The Netherlands, 2011; pp. 241–289. [Google Scholar]
- McCabe, G.M.; Fedigan, L.M. Effects of Reproductive Status on Energy Intake, Ingestion Rates, and Dietary Composition of Female Cebus capucinus at Santa Rosa, Costa Rica. Int. J. Primatol. 2007, 28, 837–851. [Google Scholar] [CrossRef]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef]
- Hicks, A.L.; Lee, K.J.; Couto-Rodriguez, M.; Patel, J.; Sinha, R.; Guo, C.; Olson, S.H.; Seimon, A.; Seimon, T.A.; Ondzie, A.U.; et al. Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat. Commun. 2018, 9, 1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeney, K.M.; Finlay, B.B. Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut. Curr. Opin. Microbiol. 2011, 14, 92–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Collado, M.C.; Isolauri, E.; Laitinen, K.; Salminen, S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 2008, 88, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Wang, X.; Bernstein, S.; Huffman, M.A.; Xia, D.P.; Gu, Z.; Chen, R.; Sheeran, L.K.; Wagner, R.S.; Li, J. Marked variation between winter and spring gut microbiota in free-ranging Tibetan Macaques (Macaca thibetana). Sci. Rep. 2016, 6, 26035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Looney, W.J.; Narita, M.; Mühlemann, K. Stenotrophomonas maltophilia: An emerging opportunist human pathogen. Lancet Infect. Dis. 2009, 9, 312–323. [Google Scholar] [CrossRef]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Cui, K.; Qi, M.; Wang, S.; Diao, Q.; Zhang, N. Dietary energy and protein levels influenced the growth performance, ruminal morphology and fermentation and microbial diversity of lambs. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Crittenden, A.N. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 3654. [Google Scholar] [CrossRef]
- De Menezes, A.B.; Lewis, E.; Odonovan, M.; Oneill, B.F.; Clipson, N.; Doyle, E.M. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 2011, 78, 256–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharechahi, J.; Zahiri, H.S.; Noghabi, K.A.; Salekdeh, G.H. In-depth diversity analysis of the bacterial community resident in the camel rumen. Syst. Appl. Microbiol. 2015, 38, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Rombouts, J.L.; Kranendonk, E.M.M.; Regueira, A.; Weissbrodt, D.G.; Kleerebezem, R.; Van Loosdrecht, M.C.M. Selecting for lactic acid producing and utilising bacteria in anaerobic enrichment cultures. Biotechnol. Bioeng. 2020, 117, 1281–1293. [Google Scholar] [CrossRef] [PubMed]
- Parvez, S.; Malik, K.A.; Kang, S.A.; Kim, H. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006, 100, 1171–1185. [Google Scholar] [CrossRef]
- Sánchez, B.; González-Rodríguez, I.; Arboleya, S.; López, P.; Suárez, A.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M. The effects of Bifidobacterium breve on immune mediators and proteome of HT29 cells monolayers. BioMed Res. Int. 2015, 2015, 479140. [Google Scholar] [CrossRef] [Green Version]
- Rao, R.K.; Samak, G. Protection and Restitution of Gut Barrier by Probiotics: Nutritional and Clinical Implications. Curr. Nutr. Food Sci. 2013, 9, 99–107. [Google Scholar]
- Stamm, R.A.; Houghton, L.A. Nutrient Intake Values for Folate during Pregnancy and Lactation Vary Widely around the World. Nutrients 2013, 5, 3920–3947. [Google Scholar] [CrossRef] [Green Version]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Yasuda, K.; Oh, K.; Ren, B.; Tickle, T.L.; Franzosa, E.A.; Wachtman, L.M.; Miller, A.D.; Westmoreland, S.V.; Mansfield, K.G.; Vallender, E.J. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 2015, 17, 385–391. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, B.; Xu, X.; Xia, Y.; Cheng, Y.; Mao, S.; Xiang, X.; Xia, D.; Wang, X.; Li, J. Variation of Gut Microbiome in Free-Ranging Female Tibetan Macaques (Macaca thibetana) across Different Reproductive States. Animals 2021, 11, 39. https://doi.org/10.3390/ani11010039
Sun B, Xu X, Xia Y, Cheng Y, Mao S, Xiang X, Xia D, Wang X, Li J. Variation of Gut Microbiome in Free-Ranging Female Tibetan Macaques (Macaca thibetana) across Different Reproductive States. Animals. 2021; 11(1):39. https://doi.org/10.3390/ani11010039
Chicago/Turabian StyleSun, Binghua, Xiaojuan Xu, Yingna Xia, Yumei Cheng, Shuxin Mao, Xingjia Xiang, Dongpo Xia, Xi Wang, and Jinhua Li. 2021. "Variation of Gut Microbiome in Free-Ranging Female Tibetan Macaques (Macaca thibetana) across Different Reproductive States" Animals 11, no. 1: 39. https://doi.org/10.3390/ani11010039
APA StyleSun, B., Xu, X., Xia, Y., Cheng, Y., Mao, S., Xiang, X., Xia, D., Wang, X., & Li, J. (2021). Variation of Gut Microbiome in Free-Ranging Female Tibetan Macaques (Macaca thibetana) across Different Reproductive States. Animals, 11(1), 39. https://doi.org/10.3390/ani11010039





