Increasing Fat Deposition Via Upregulates the Transcription of Peroxisome Proliferator-Activated Receptor Gamma in Native Crossbred Chickens
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Birds and Rearing Condition
2.2. Slaughtering, Fat Deposition Data, and Tissue Collection
2.3. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.4. Statistical Analysis
3. Results
3.1. Carcass Fat Deposition among Various Genetic Background of Chickens
3.2. Differentiation of PPARA mRNA Expression in Various Breed of Chickens
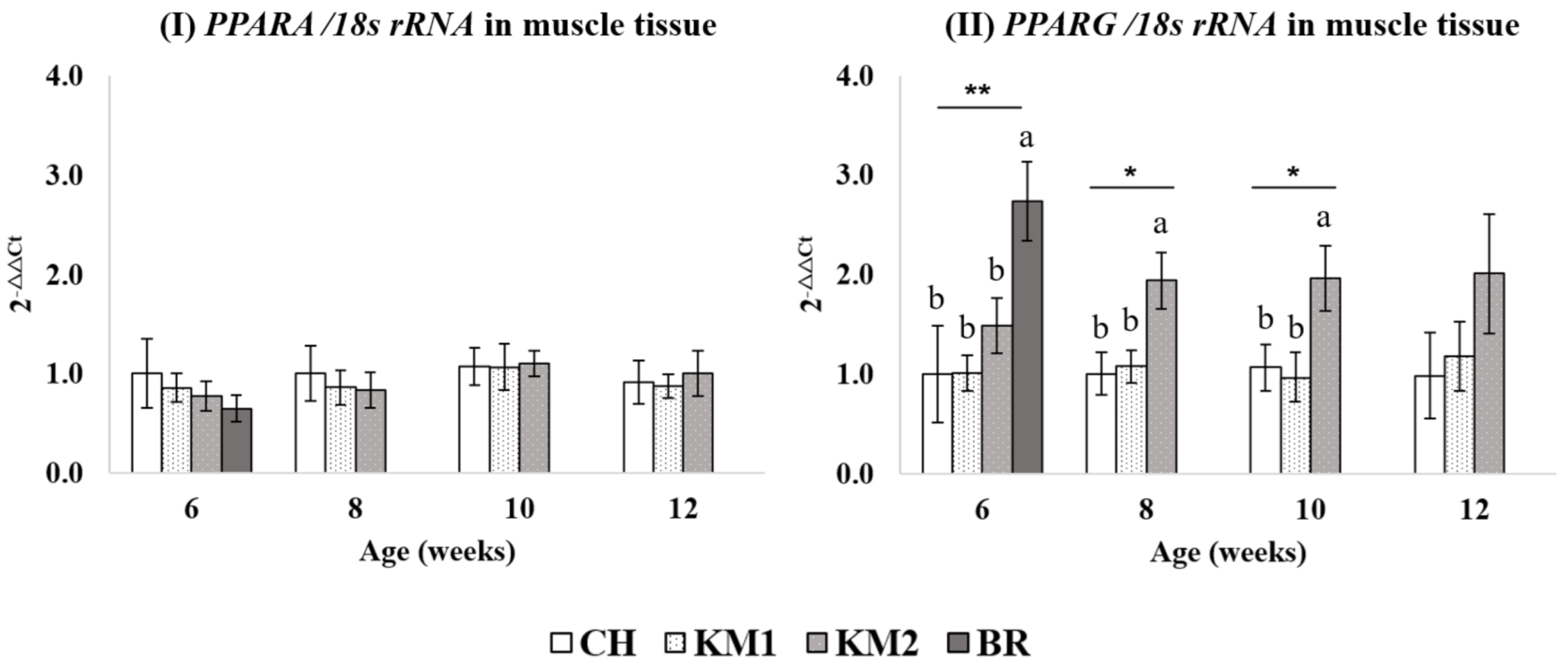
3.3. Differentiation of PPARG mRNA Expression in Various Breed of Chickens
3.4. Correlation between PPARs Expression and Fat Deposition in Slaughtering Trait
4. Discussion
4.1. Impact of Crossbreeding of Thai Native Chickens on Growth
4.2. PPARs Transcription Factors Regulating Cellular Lipid Metabolism
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaturasitha, S.; Chaiwang, N.; Kreuzer, M. Thai native chicken meat: An option to meet the demands for specific meat quality by certain groups of consumers; a review. Anim. Prod. Sci. 2017, 57, 1582. [Google Scholar] [CrossRef]
- Padhi, M.K. Importance of Indigenous Breeds of Chicken for Rural Economy and Their Improvements for Higher Production Performance. Scientifica 2016, 2016, 2604685. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Zheng, M.; Zhao, G.; Liu, R.; Wen, J. Identification of differentially expressed genes and pathways for intramuscular fat metabolism between breast and thigh tissues of chickens. BMC Genom. 2018, 19, 55. [Google Scholar] [CrossRef]
- Meng, H.; Li, H.; Zhao, J.G.; Gu, Z.L. Differential expression of peroxisome proliferator-activated receptors alpha and gamma gene in various chicken tissues. Domest. Anim. 2005, 28, 105–110. [Google Scholar] [CrossRef]
- Spiegelman, B.M. Peroxisome proliferator-activated receptor gamma: A key regulator of adipogenesis and systemic insulin sensitivity. Eur. J. Med. Res. 1997, 2, 457–464. [Google Scholar]
- Wang, H.-B.; Li, H.; Wang, Q.-G.; Zhang, X.-Y.; Wang, S.-Z.; Wang, Y.-X.; Wang, X.-P. Profiling of chicken adipose tissue gene expression by genome array. BMC Genom. 2007, 8, 193. [Google Scholar] [CrossRef] [Green Version]
- Royan, M.; Navidshad, B. Peroxisome proliferator-activated receptor gamma (PPARγ), a key regulatory gene of lipid metabolism in chicken. Worlds Poult. Sci. J. 2016, 72, 773–784. [Google Scholar] [CrossRef]
- Sato, K.; Abe, H.; Kono, T.; Yamazaki, M.; Nakashima, K.; Kamada, T.; Akiba, Y. Changes in peroxisome proliferator-activated receptor gamma gene expression of chicken abdominal adipose tissue with different age, sex and genotype. Anim. Sci. J. 2009, 80, 322–327. [Google Scholar] [CrossRef]
- Dreyer, C.; Keller, H.; Mahfoudi, A.; Laudet, V.; Krey, G.; Wahli, W. Positive regulation of the peroxisomal β-oxidation pathway by fatty acids through activation of peroxisome proliferator-activated receptors (PPAR). Biol. Cell 1993, 77, 67–76. [Google Scholar] [CrossRef]
- AOAC International. Official Methods of Analysis (AOAC), 19th ed.; Latimed, G., Ed.; Association of Official Analytical Chemist: Gaithersburg, MD, USA, 2012. [Google Scholar]
- Ramiah, S.K.; Meng, G.Y.; Sheau Wei, T.; Swee Keong, Y.; Ebrahimi, M. Dietary Conjugated Linoleic Acid Supplementation Leads to Downregulation of PPAR Transcription in Broiler Chickens and Reduction of Adipocyte Cellularity. PPAR Res. 2014, 2014, 137652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenwick, M.A.; Fitzpatrick, R.; Kenny, D.A.; Diskin, M.G.; Patton, J.; Murphy, J.J.; Wathes, D.C. Interrelationships between negative energy balance (NEB) and IGF regulation in liver of lactating dairy cows. Domest. Anim. 2008, 34, 31–44. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yin, H.D.; Gilbert, E.R.; Chen, S.Y.; Wang, Y.; Zhang, Z.C.; Zhao, X.L.; Zhang, Y.; Zhu, Q. Effect of Hybridization on Carcass Traits and Meat Quality of Erlang Mountainous Chickens. Asian-Australas. J. Anim. Sci. 2013, 26, 1504–1510. [Google Scholar] [CrossRef] [Green Version]
- Zerehdaran, S.; Vereijken, A.L.J.; Van Arendonk, J.A.M.; Van der Waaijt, E.H. Estimation of genetic parameters for fat deposition and carcass traits in broilers. Poult. Sci. 2004, 83, 521–525. [Google Scholar] [CrossRef]
- Hocquette, J.F.; Gondret, F.; Baéza, E.; Médale, F.; Jurie, C.; Pethick, D.W. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2010, 4, 303–319. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.P.; Chen, J.L.; Zheng, M.Q.; Wen, J.; Zhang, Y. Correlated responses to selection for increased intramuscular fat in a Chinese quality chicken line. Poult. Sci. 2007, 86, 2309–2314. [Google Scholar] [CrossRef]
- Jaturasitha, S.; Kayan, A.; Wicke, M. Carcass and meat characteristics of male chickens between Thai indigenous compared with improved layer breeds and their crossbred. Arch. Anim. Breed. 2008, 51, 283–294. [Google Scholar] [CrossRef]
- Wang, G.; Kim, W.K.; Cline, M.A.; Gilbert, E.R. Factors affecting adipose tissue development in chickens: A review. Poult. Sci. 2017, 96, 3687–3699. [Google Scholar] [CrossRef]
- Hausman, G.J.; Dodson, M.V.; Ajuwon, K.; Azain, M.; Barnes, K.M.; Guan, L.L.; Jiang, Z.; Poulos, S.P.; Sainz, R.D.; Smith, S.; et al. Board-invited review: The biology and regulation of preadipocytes and adipocytes in meat animals. J. Anim. Sci. 2009, 87, 1218–1246. [Google Scholar] [CrossRef] [Green Version]
- Grabacka, M.; Pierzchalska, M.; Dean, M.; Reiss, K. Regulation of Ketone Body Metabolism and the Role of PPARα. Int. J. Mol. Sci. 2016, 17, 2093. [Google Scholar] [CrossRef] [Green Version]
- Takada, I.; Kobayashi, M. Structural Features and Transcriptional Activity of Chicken PPARs (α, β, and γ). PPAR Res. 2013, 2013, 186312. [Google Scholar] [CrossRef] [Green Version]
- Meertens, L.M.; Miyata, K.S.; Cechetto, J.D.; Rachubinski, R.A.; Capone, J.P. A mitochondrial ketogenic enzyme regulates its gene expression by association with the nuclear hormone receptor PPARalpha. EMBO J. 1998, 17, 6972–6978. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Lin, S.; Mu, H.; Tang, X.; Ou, Y.; Chen, J.; Ma, Y.; Li, Y. Analysis of Differentially Expressed Genes and Signaling Pathways Related to Intramuscular Fat Deposition in Skeletal Muscle of Sex-Linked Dwarf Chickens. Biomed. Res. Int. 2014, 2014, 724274. [Google Scholar] [CrossRef]
- Matsubara, Y.; Sato, K.; Ishii, H.; Akiba, Y. Changes in mRNA expression of regulatory factors involved in adipocyte differentiation during fatty acid induced adipogenesis in chicken. Comp. Biochem. Physiol. Part. A Mol. Integr. Physiol. 2005, 141, 108–115. [Google Scholar] [CrossRef]
- Wang, Y.; Mu, Y.; Li, H.; Ding, N.; Wang, Q.; Wang, Y.; Wang, S.; Wang, N. Peroxisome Proliferator-Activated Receptor-γ Gene: A Key Regulator of Adipocyte Differentiation in Chickens. Poult. Sci. 2008, 87, 226–232. [Google Scholar] [CrossRef]
- Tunim, S.; Phasuk, Y.; Aggrey, S.E.; Duangjinda, M. Gene expression of fatty acid binding protein genes and its relationship with fat deposition of Thai native crossbreed chickens. Asian-Australas. J. Anim. Sci. 2020. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Niu, J.; Geng, Z. Gene expression and plasma lipid content in relation to intramuscular fat in Chinese indigenous Wuhua chicken. J. Appl. Poult. Res. 2017, 26, 391–400. [Google Scholar] [CrossRef]
- Wang, X.G.; Yu, J.F.; Zhang, Y.; Gong, D.Q.; Gu, Z.L. Identification and characterization of microRNA from chicken adipose tissue and skeletal muscle. Poult. Sci. 2012, 91, 139–149. [Google Scholar] [CrossRef]
- Zhang, M.; Li, D.-H.; Li, F.; Sun, J.-W.; Jiang, R.-R.; Li, Z.-J.; Han, R.-L.; Li, G.-X.; Liu, X.-J.; Kang, X.-T.; et al. Integrated Analysis of MiRNA and Genes Associated with Meat Quality Reveals that Gga-MiR-140-5p Affects Intramuscular Fat Deposition in Chickens. Cell Physiol. Biochem. 2018, 46, 2421–2433. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Shan, Y.; Ji, G.; Ju, X.; Tu, Y.; Sheng, Z.; Xie, J.; Zou, J.; Shu, J. miRNA–mRNA network regulation in the skeletal muscle fiber phenotype of chickens revealed by integrated analysis of miRNAome and transcriptome. Sci. Rep. 2020, 10, 10619. [Google Scholar] [CrossRef]



| Genes | Sequences | Product (bp) | TM | Sources |
|---|---|---|---|---|
| PPARA | F: 5- AGGCCAAGTTGAAAGCAGA-3 R: 5-GTCTTCTCTGCCATGCACAA-3 | 217 | 58 | [11] |
| PPARG | F: 5-GACCTTAATTGTCGCATCCAT-3 R: 5-CGGGAAGGACTTTATGTATGA-3 | 237 | 56 | [11] |
| 18S rRNA | F: 5-CGGCGACGACCCATTCGAAC-3 R: 5-GAATCGAACCCTGATTCCCCGTC-3 | 99 | 62 | [12] |
| Fat Deposition Traits (%) | Adipose Tissue | Muscular Tissue | ||
|---|---|---|---|---|
| PPARA (n = 102) | PPARG (n = 102) | PPARA (n = 102) | PPARG (n = 102) | |
| Abdominal fat (n = 258) | −0.02 | 0.38 ** | −0.02 | 0.37 ** |
| Subcutaneous fat (n = 260) | −0.18 | 0.10 | 0.01 | 0.28 ** |
| Skin fat (n = 104) | −0.26 * | 0.32 ** | −0.06 | 0.20 * |
| Breast intramuscular fat (n = 104) | −0.23 * | 0.22 * | −0.04 | 0.39 ** |
| Thigh intramuscular fat (n = 104) | −0.28 ** | 0.36 ** | −0.04 | 0.46 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tunim, S.; Phasuk, Y.; Aggrey, S.E.; Duangjinda, M. Increasing Fat Deposition Via Upregulates the Transcription of Peroxisome Proliferator-Activated Receptor Gamma in Native Crossbred Chickens. Animals 2021, 11, 90. https://doi.org/10.3390/ani11010090
Tunim S, Phasuk Y, Aggrey SE, Duangjinda M. Increasing Fat Deposition Via Upregulates the Transcription of Peroxisome Proliferator-Activated Receptor Gamma in Native Crossbred Chickens. Animals. 2021; 11(1):90. https://doi.org/10.3390/ani11010090
Chicago/Turabian StyleTunim, Supanon, Yupin Phasuk, Samuel E. Aggrey, and Monchai Duangjinda. 2021. "Increasing Fat Deposition Via Upregulates the Transcription of Peroxisome Proliferator-Activated Receptor Gamma in Native Crossbred Chickens" Animals 11, no. 1: 90. https://doi.org/10.3390/ani11010090





