Identification of microRNAs in Silver Carp (Hypophthalmichthys molitrix) Response to Hypoxia Stress
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Experimental Fish and Sample Collection
2.3. Total RNA Extraction, Library Construction and Small RNA Sequencing
2.4. Identification and Analysis of Differentially Expressed miRNAs
2.5. Real-Time PCR Validation
3. Results
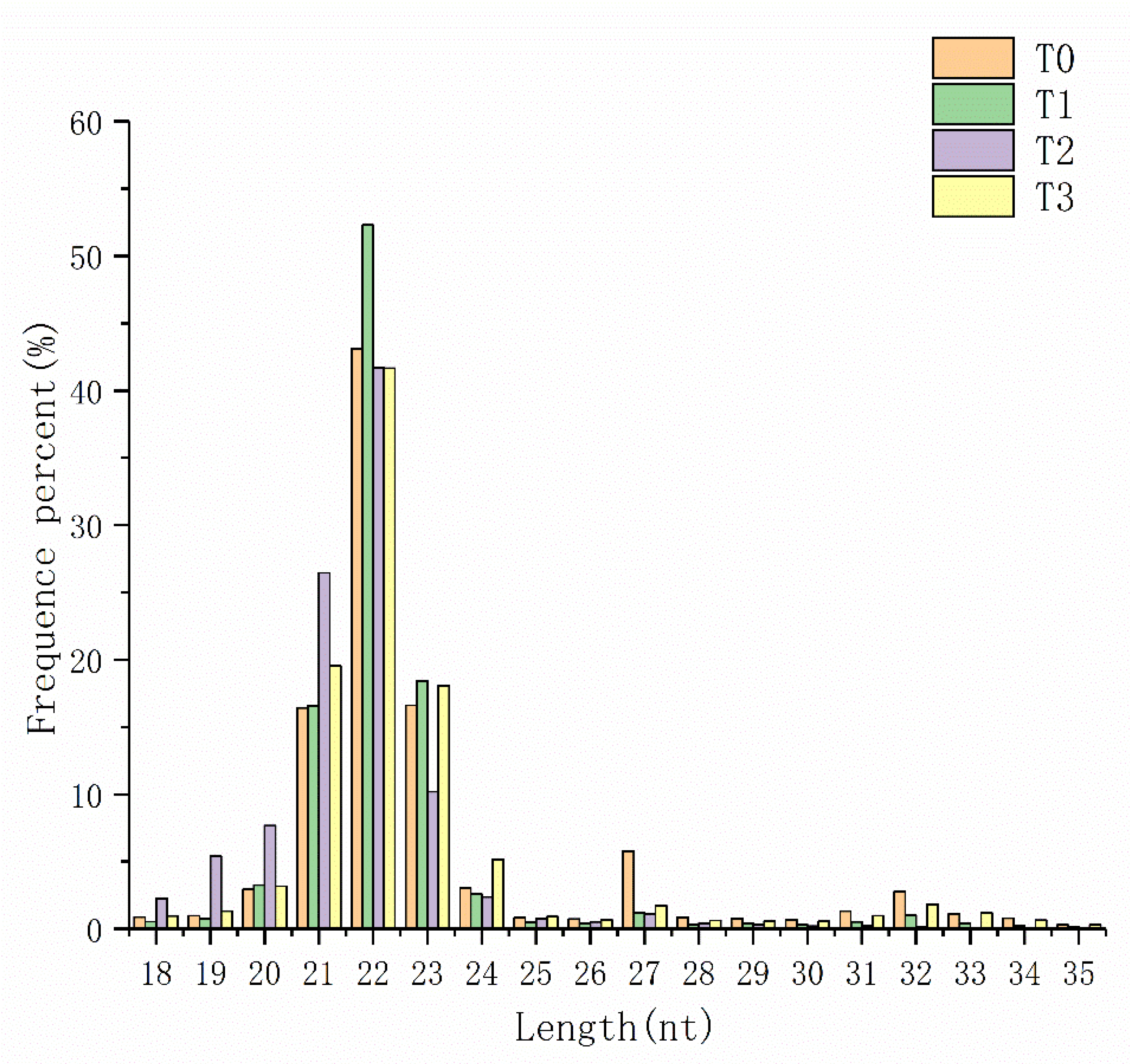
3.1. Analysis of miRNA Library Sequencing Data
3.2. Differential Expression Analysis of miRNAs
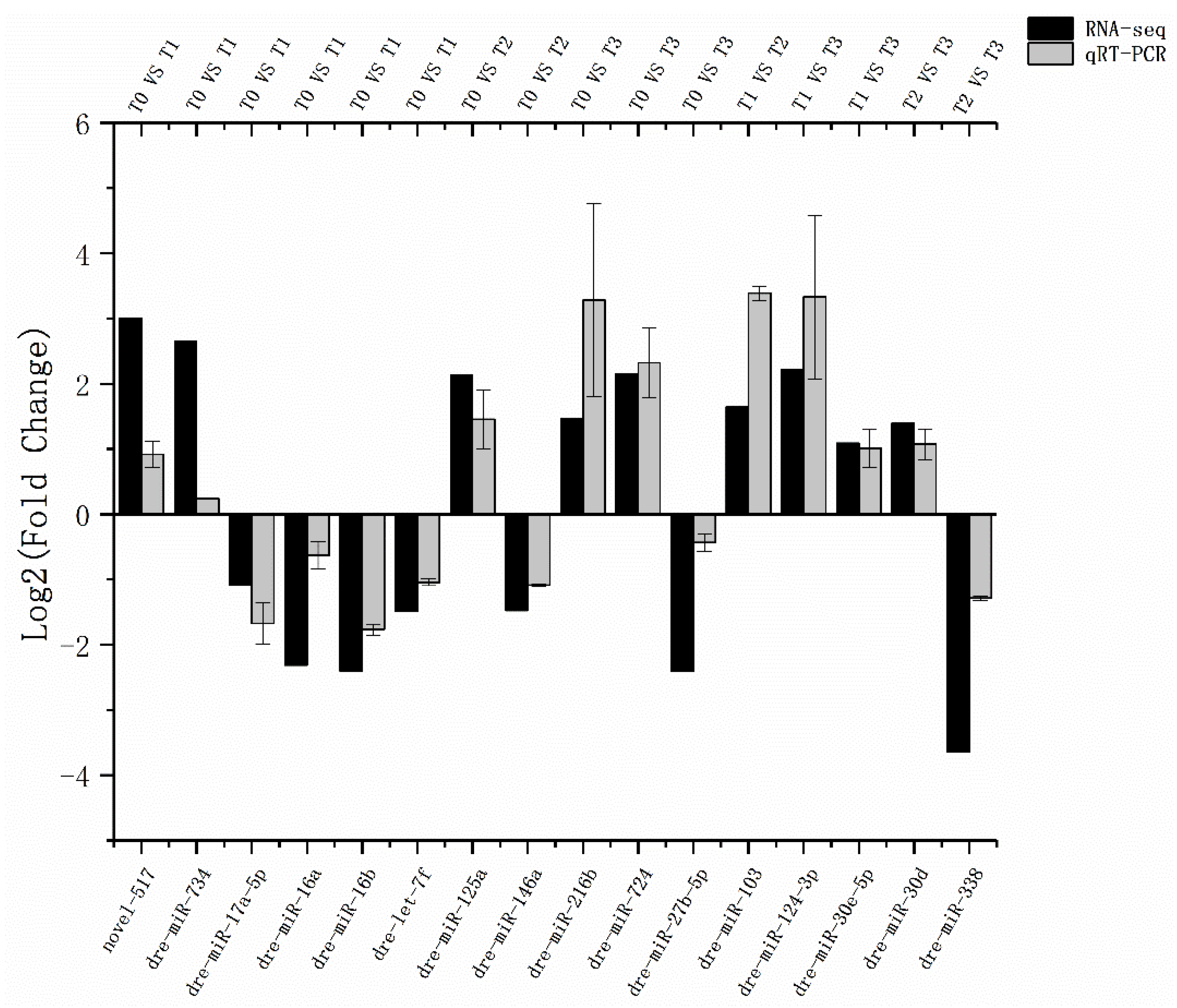
3.3. Validation of Selected miRNAs by Real-Time PCR
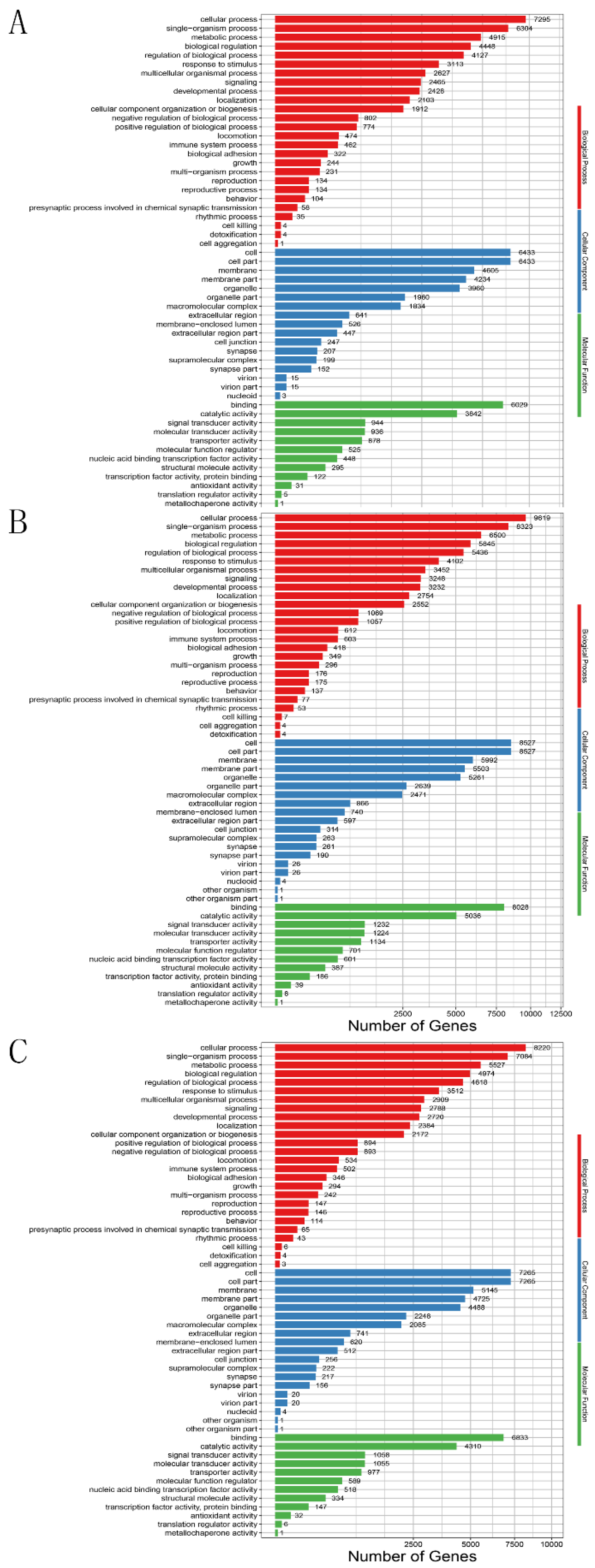
3.4. Target Gene Prediction and Functional Annotation of Differentially Expressed miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Rudolf, S.S.W. Hypoxia: From molecular responses to ecosystem responses. Mar. Pollut. Bull. 2002, 45, 35–45. [Google Scholar]
- Pollock, M.S.; Clarke, L.M.J.; Dubé, M.G. The effects of hypoxia on fishes: From ecological relevance to physiological effects. Environ. Rev. 2007, 15, 1–14. [Google Scholar] [CrossRef]
- Ruan, W.; Ji, W.W.; Zheng, L.; Yue, D.D.; Fang, H. On hypoxia stress in fish and its nutritional regulation and response. Mar. Fish. 2020, 42, 751–761. [Google Scholar]
- Thomas, O’.C.; David, W. Linking hypoxia to shrimp catch in the northern Gulf of Mexico. Mar. Pollut. Bull. 2007, 54, 460–463. [Google Scholar]
- Raquel, V.S.; Carlos, M.D. Thresholds of hypoxia for marine biodiversity. Proc. Natl. Acad. Sci. USA 2008, 105, 15452–15457. [Google Scholar]
- Ding, C.Y.; Hu, L.S.; Li, Y.; Xue, Y.; Li, H.; Wu, R.H.; Liu, E.X.; Li, X.J. Effects of hypoxia stress on cardiomyocyte apoptosis and the control for Bax, Bcl-2 expressions in Hypophthalmichthys molitri. Free Fish. 2018, 48, 10–15. [Google Scholar]
- Zhao, J.K.; Liang, H.W.; Zou, G.W.; Wang, H.L.; Li, Z. Influence of hypoxic stress on apoptosis of hepatocyte and brain cells of silver carp (Hypophthalmichthys molitri). J. Northwest A&F Univ. (Nat. Sci. Ed.) 2016, 44, 34–38. [Google Scholar]
- Huang, C.X.; Chen, N.; Wu, X.J.; Huang, C.H.; He, Y.; Tang, R.; Wang, W.M.; Wang, H.L. The zebrafish miR-462/miR-731 cluster is induced under hypoxic stress via hypoxia-inducible factor 1a and functions in cellular adaptations. FASEB J. 2015, 29, 4901–4913. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.A.; Luo, Y.T.; Xu, D.P.; Yang, X.W.; Wang, X.H. Relationship between genetic risk and stock enhancement of the silver carp (Hypophthalmichthys molitrix) in the Yangtze River. Fish. Res. 2021, 235, 105829. [Google Scholar] [CrossRef]
- Wang, X.; Wang, B.L.; Xia, C.X.; Bi, Y.H.; Chen, L.; Hu, Z.Y. The predator effects of sliver carp on different algal species inside aquarium. Acta Hydrobiol. Sinica 2015, 39, 940–947. [Google Scholar]
- Qiang, J.; He, J.; Tao, Y.F.; Bao, J.W.; Zhu, J.H.; Xu, P. Hypoxia-induced miR-92a regulates p53 signaling pathway and apoptosis by targeting calcium-sensing receptor in genetically improved farmed tilapia (Oreochromis niloticus). PLoS ONE 2020, 15, 0238897. [Google Scholar] [CrossRef]
- Xie, J.; Wang, Q. Research progress of environmental stress and physiological regulation mechanism in aquatic animal during keep live transportation. Food. Sci. 2020, 42, 319–325. [Google Scholar]
- Minju, H.V.; Narry, K. Regulation of microrna biogenesis. Nat. Rev. Mol. Cell. Bio. 2014, 15, 509–524. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402. [Google Scholar] [CrossRef]
- Ana, K.; Maria, B.; Sam, G.J. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, 155–162. [Google Scholar]
- Eric, A.M. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005, 15, 563–568. [Google Scholar]
- Sun, J.L.; Zhao, L.L.; He, K.; Liu, Q.; Yang, S. MiRNA-mRNA integration analysis reveals the regulatory roles of miRNAs in the metabolism of largemouth bass (Micropterus salmoides) livers during acute hypoxic stress. Aquaculture 2020, 526, 1–10. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, C.D.; Yan, B.; Zhao, J.L.; Wang, Z.H. miRNA-directed regulation of VEGF in tilapia under hypoxia condition. Biochem. Biophys. Res. Commun. 2014, 10, 183. [Google Scholar] [CrossRef]
- Huang, Y.; Gong, W.B.; Xiong, J.L.; Gao, X.C.; Ren, H.T. Discovery and characterization of conserved and novel microRNAs from blunt snout bream (Megalobrama amblycephala) by deep sequencing. Gene 2018, 654, 57–63. [Google Scholar] [CrossRef]
- Li, C.; Xu, D.X. Understanding microRNAs regulation in heat shock response in the sea cucumber Apostichopus japonicus. Fish Shellfish Immun. 2018, 81, 214–220. [Google Scholar] [CrossRef]
- Wang, W.; Zhong, P.; Yi, J.Q.; Xu, A.X.; Lin, W.Y.; Guo, Z.C.; Wang, C.G.; Sun, C.B.; Chan, S. Potential role for microRNA in facilitating physiological adaptation to hypoxia in the Pacific whiteleg shrimp Litopenaeus vannamei. Fish Shellfish Immun. 2018, 84, 361–369. [Google Scholar] [CrossRef]
- Chen, H.; Xin, L.S.; Song, X.R.; Wang, L.; Wang, W.L.; Liu, Z.J.; Zhang, H.; Wang, L.G.; Zhou, Z.; Qiu, L.M.; et al. A norepinephrine-responsive miRNA directly promotes CgHSP90AA1 expression in oyster haemocytes during desiccation. Fish. Shellfish Immun. 2017, 64, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Rosalind, C.L.; Rhonda, L.F.; Victor, A. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar]
- Nallamshetty, S.; Chan, S.Y.; Loscalzo, J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radic. Biol. Med. 2013, 64, 20–30. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.Z.; Yang, C.X.; Ma, W.W.; Wang, Y.; Feng, M.X.; Dong, W.G.; Wang, X.; Zhang, M.; Dong, J.; Gao, Y.N.; et al. A review: Progress of microRNA research on adversity to fish. Fish. Sci. 2020, 39, 771–779. [Google Scholar]
- Zhao, C.; Fan, S.A.; Qiu, L.H. Identification of MicroRNAs and Their Target Genes Associated with Ovarian Development in Black Tiger Shrimp (Penaeus monodon) Using High-Throughput Sequencing. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Karen, L.; Keng, P.L.; Jessie, Y.J.B.; Na, Z.; Anna, T.; Amy, T.; Jing, W.L.; Si, L.; Richard, Y.C.K.; Wing, Y.L.; et al. Identification and expression profiling of microRNAs in the brain, liver and gonads of marine medaka (Oryzias melastigma) and in response to hypoxia. PLoS ONE 2014, 9, e110698. [Google Scholar]
- Zhong, T.; Wang, C.; Hu, J.T.; Chen, X.Y.; Niu, L.L.; Zhan, S.Y.; Wang, L.J.; Guo, Z.; Cao, J.X.; Li, L.; et al. Comparison of MicroRNA Transcriptomes Reveals the Association between MiR-148a-3p Expression and Rumen Development in Goats. Animals 2020, 10, 1951. [Google Scholar] [CrossRef]
- Hou, Z.B.; Xie, L.; Yu, L.X.; Qian, X.P.; Liu, B.R. MicroRNA-146a is down-regulated in gastric cancer and regulates cell proliferation and apoptosis. Med. Oncol. 2012, 29, 886–892. [Google Scholar] [CrossRef]
- Peschiaroli, A.; Giacobbe, A.; Formosa, A.; Markert, E.K.; Bongiorno, B.L.; Levine, A.J.; Candi, E.; D'Alessandro, A.; Zolla, L.; Finazzi, A.; et al. MiR-143 regulates hexokinase 2 expression in cancer cells. Oncogene 2013, 32, 797–802. [Google Scholar] [CrossRef]
- Hall, J.R.; Short, C.E.; Petersen, L.H.; Stacey, J.; Gamperl, A.K.; Driedzic, W.R. Expression levels of genes associated with oxygen utilization, glucose transport and glucose phosphorylation in hypoxia exposed Atlantic cod (Gadus morhua). Comp. Biochem. Phys. D 2009, 4, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.T.; Sun, J.J.; Liu, Y.; Zheng, X.G.; Yang, H.B. Effect of mir-125a-5p targeting scarb1 gene on hypoxia/reoxygenation injury of cardiomyocytes and its mechanism. Chin. J. Med. Genet. 2020, 37, 980–986. [Google Scholar]
- Sikorska, M.; Siwek, M.; Slawinska, A.; Dunislawska, A. MiRNA Profiling in the Chicken Liver under the Influence of Early Microbiota Stimulation with Probiotic, Prebiotic, and Synbiotic. Genes 2021, 12, 685. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Li, H.L.; Xu, B.H.; Liu, Z.M.; Zhang, X.F.; Xia, S.H. Effect of miR-21 on hypoxia-reoxygenation injury in hepatocellular. Shandong Med. J. 2018, 58, 31–34. [Google Scholar]
- Wei, D.H. Research progress of the effect of miRNA on fat metabolism in sheep under acute cold stress. Mod. J. Anim. Husb. Vet. Med. 2020, 10, 55–60. [Google Scholar]
- Zhang, J.; Luo, H.; Xiong, Z.B.; Wan, K.; Liao, Q.F.; He, H. High-throughput sequencing reveals biofluid exosomal miRNAs associated with immunity in pigs. Biosci. Biotech. Biochem. 2020, 84, 53–62. [Google Scholar] [CrossRef]
- Xu, J.S.; Hu, H.T.; Li, H.L.; Chang, S.W. The Role of miRNAs in Immune Cell Development, Immune Cell Activation, and Tumor Immunity: With a Focus on Macrophages and Natural Killer Cells. Cells 2019, 8, 1140. [Google Scholar] [CrossRef] [Green Version]

| miRNAs | Primer | Primer Sequence (5′-3′) |
|---|---|---|
| dre-let-7f | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACTAT |
| F | GCGCGCTGAGGTAGTAGATTGT | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-125a | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACAGG |
| F | GCGCTCCCTGAGACCCTTAA | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-216b | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACAG |
| F | GCGCGCTAATCTCTGCAGGCAA | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-724 | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACAGT |
| F | GCGCGCTTAAAGGGAATTTGCG | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-103 | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCATAG |
| F | GCGCAGCAGCATTGTACAGGG | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-146a | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCATCT |
| F | GCGCGCTGAGAACTGAATTCCAT | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-27b-5p | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGTTCA |
| F | GCGCGAGAGCTTAGCTGATTGG | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-124-3p | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTGGCA |
| F | GCGCTAAGGCACGCGGTGAA | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-30e-5p | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTTCCA |
| F | GCGCGCTGTAAACATCCTTGAC | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-338 | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAACAA |
| F | GCGCGCTCCAGCATCAGTGATT | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-30d | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTTCCA |
| F | GCGCTGTAAACATCCCCGAC | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-734 | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGGTAC |
| F | GCGCGCGTAAATGCTGCAGAATC | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-17a-5p | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTACCTG |
| F | GCGCGCCAAAGTGCTTACAGTG | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-16a | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACCAA |
| F | GCGCGCTAGCAGCACGTAAATA | |
| R | GCAGGGTCCGAGGTATTC | |
| dre-miR-16b | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTCCAA |
| F | GCGCGCTAGCAGCACGTAAATA | |
| R | GCAGGGTCCGAGGTATTC | |
| novel-517 | Loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGCTCA |
| F | GCGCGCACCTACACTGTCTAC | |
| R | GCAGGGTCCGAGGTATTC | |
| U6 | Loop | AAAACAGCAATATGGAGCGC |
| F | TGCTCGCTACGGTGGCACA | |
| R | AAAACAGCAATATGGAGCGC |
| Items | T0 | T1 | T2 | T3 |
|---|---|---|---|---|
| Total reads | 26,475,225 | 26,143,905 | 22,941,194 | 28,984,723 |
| Clean reads | 25,558,042 | 25,542,062 | 19,466,537 | 28,030,238 |
| Q20 | 99.65% | 99.65% | 99.64% | 99.62% |
| Q30 | 98.98% | 98.97% | 98.94% | 98.88% |
| GC content | 48.38% | 47.87% | 49.12% | 48.28% |
| miRNAs | miRNA Sequence (5′-3′) | T0 | T1 | T2 | T3 |
|---|---|---|---|---|---|
| dre-miR-100-5p | AACCCGUAGAUCCGAACUUGUG | 2,167,459 | 1,924,641 | 1,089,212 | 1,969,635 |
| dre-miR-143 | UGAGAUGAAGCACUGUAGCUC | 1,113,425 | 641,659 | 2,373,465 | 1,570,961 |
| dre-miR-101a | UACAGUACUGUGAUAACUGAAG | 688,497 | 1,472,818 | 484,507 | 396,814 |
| dre-miR-146a | UGAGAACUGAAUUCCAUAGAUGG | 167,716 | 70,684 | 248,036 | 193,016 |
| dre-miR-21 | UAGCUUAUCAGACUGGUGUUGGC | 416,516 | 318,134 | 327,475 | 347,802 |
| dre-miR-22a-3p | AAGCUGCCAGCUGAAGAACUGU | 757,583 | 492,558 | 1,363,640 | 664,397 |
| dre-miR-26a-5p | UUCAAGUAAUCCAGGAUAGGCU | 670,590 | 490,815 | 706,568 | 666,125 |
| dre-miR-30d | UGUAAACAUCCCCGACUGGAAG | 190,391 | 226,262 | 352,041 | 202,079 |
| dre-miR-99 | AACCCGUAGAUCCGAUCUUGUG | 613,288 | 668,863 | 1,041,263 | 667,971 |
| dre-let-7e | UGAGGUAGUAGAUUGAAUAGUU | 216,109 | 443,662 | 160,268 | 439,320 |
| dre-miR-30e-5p | UGUAAACAUCCUUGACUGGAAG | 92,054 | 103,939 | 119,573 | 48,008 |
| dre-miR-451 | AAACCGUUACCAUUACUGAGUU | 31,188 | 16,482 | 36,582 | 23,333 |
| dre-miR-27b-3p | UUCACAGUGGCUAAGUUCUGCA | 88,506 | 40,150 | 223,389 | 84,618 |
| Types | T0 | T0 (Percent) | T1 | T1 (Percent) | T2 | T2 (Percent) | T3 | T3 (Percent) |
|---|---|---|---|---|---|---|---|---|
| total | 22,823,393 | 100.00% | 24,310,475 | 100.00% | 17,914,838 | 100.00% | 25,405,520 | 100.00% |
| known_miRNA | 16,902,843 | 74.06% | 19,366,521 | 79.66% | 13,751,148 | 76.76% | 18,336,438 | 72.18% |
| rRNA | 408,035 | 1.79% | 201,337 | 0.83% | 545,212 | 3.04% | 981,912 | 3.86% |
| tRNA | 2 | 0.00% | 0 | 0.00% | 8 | 0.00% | 2 | 0.00% |
| snRNA | 27,663 | 0.12% | 19,041 | 0.08% | 11,027 | 0.06% | 17,458 | 0.07% |
| snoRNA | 11,148 | 0.05% | 8611 | 0.04% | 36,446 | 0.20% | 22,629 | 0.09% |
| repeat | 1,506,607 | 6.60% | 532,386 | 2.19% | 154,534 | 0.86% | 971,529 | 3.82% |
| novel_miRNA | 132,493 | 0.58% | 90,735 | 0.37% | 94,536 | 0.53% | 121,857 | 0.48% |
| exon: + | 141,310 | 0.62% | 60,768 | 0.25% | 336,910 | 1.88% | 170,306 | 0.67% |
| exon: − | 112,890 | 0.49% | 44,975 | 0.19% | 272,386 | 1.52% | 138,940 | 0.55% |
| intron: + | 164,073 | 0.72% | 103,326 | 0.43% | 85,369 | 0.48% | 157,480 | 0.62% |
| intron: − | 89,759 | 0.39% | 70,626 | 0.29% | 50,486 | 0.28% | 112,331 | 0.44% |
| other | 3,326,570 | 14.58% | 3,812,149 | 15.68% | 2,576,776 | 14.38% | 4,374,638 | 17.22% |
| UP | T0 | T1 | T2 | T3 | |
|---|---|---|---|---|---|
| DOWN | |||||
| T0 | 75 | 210 | 104 | ||
| T1 | 55 | 183 | 100 | ||
| T2 | 112 | 152 | 144 | ||
| T3 | 60 | 95 | 186 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Li, X.; Sha, H.; Luo, X.; Zou, G.; Liang, H. Identification of microRNAs in Silver Carp (Hypophthalmichthys molitrix) Response to Hypoxia Stress. Animals 2021, 11, 2917. https://doi.org/10.3390/ani11102917
Wang Q, Li X, Sha H, Luo X, Zou G, Liang H. Identification of microRNAs in Silver Carp (Hypophthalmichthys molitrix) Response to Hypoxia Stress. Animals. 2021; 11(10):2917. https://doi.org/10.3390/ani11102917
Chicago/Turabian StyleWang, Qiaoxin, Xiaohui Li, Hang Sha, Xiangzhong Luo, Guiwei Zou, and Hongwei Liang. 2021. "Identification of microRNAs in Silver Carp (Hypophthalmichthys molitrix) Response to Hypoxia Stress" Animals 11, no. 10: 2917. https://doi.org/10.3390/ani11102917
APA StyleWang, Q., Li, X., Sha, H., Luo, X., Zou, G., & Liang, H. (2021). Identification of microRNAs in Silver Carp (Hypophthalmichthys molitrix) Response to Hypoxia Stress. Animals, 11(10), 2917. https://doi.org/10.3390/ani11102917





