Tissue and Temperature-Specific RNA-Seq Analysis Reveals Genomic Versatility and Adaptive Potential in Wild Sea Turtle Hatchlings (Caretta caretta)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Hatchling Sample Collection and RNA Extraction
2.2. De Novo Transcriptome Assembly
2.3. Differential Gene Expression and Enrichment Analyses
3. Results
Gene Ontology Enrichment and Functional Annotation Clustering
4. Discussion
Biological Significance of Differentially Expressed Genes among Female and Male Loggerheads
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FDR | False discovery rate |
| GO | Gene Ontology |
| TSD | Temperature-dependent sex determination |
References
- Bowen, B.W.; Karl, S.A. Population genetics and phylogeography of sea turtles. Mol. Ecol. 2007, 16, 4886–4907. [Google Scholar] [CrossRef] [PubMed]
- Broderick, A.C.; Coyne, M.S.; Fuller, W.J.; Glen, F.; Godley, B.J. Fidelity and over-wintering of sea turtles. Proc. R. Soc. B. 2007, 274, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.R.; Carthy, R.R.; Ceriani, S.A. Migration routes, foraging behavior, and site fidelity of loggerhead sea turtles (Caretta caretta) satellite tracked from a globally important rookery. Mar. Biol. 2019, 166, 134. [Google Scholar] [CrossRef]
- Siegwalt, F.; Benhamou, S.; Girondot, M.; Jeantet, L.; Martin, J.; Bonola, M.; Lelong, P.; Grand, C.; Chambault, P.; Benhalilou, A.; et al. High fidelity of sea turtles to their foraging grounds revealed by satellite tracking and capture-mark-recapture: New insights for the establishment of key marine conservation areas. Biol. Conserv. 2020, 250, 108742. [Google Scholar] [CrossRef]
- Mrosovsky, N.; Kamel, S.; Rees, A.F.; Margaritoulis, D. Pivotal temperature for loggerhead turtles (Caretta caretta) from KyparissiaBay, Greece. Can. J. Zool. 2002, 80, 2118–2124. [Google Scholar] [CrossRef] [Green Version]
- Lolavar, A.; Wyneken, J. Effects of supplemental watering on loggerhead (Caretta caretta) nests and hatchlings. J. Exp. Mar. Biol. Ecol. 2021, 534, 151476. [Google Scholar] [CrossRef]
- Marco, A.; Abella, E.; Martins, S.; López, O.; Patino-Martinez, J. Female nesting behaviour affects hatchling survival and sex ratio in the loggerhead sea turtle: Implications for conservation programmes. Ethol. Ecol. Evol. 2018, 30, 141–155. [Google Scholar] [CrossRef]
- Hawkes, L.A.; Broderick, A.C.; Godfrey, M.H.; Godley, B.J. Investigating the potential impacts of climate change on a marine turtle population. Glob. Chang. Biol. 2007, 13, 923–932. [Google Scholar] [CrossRef]
- Dang, W.; Zhang, W.; Du, W.-G. Incubation temperature affects the immune function of hatchling soft-shelled turtles, Pelodiscus sinensis. Sci. Rep. 2015, 5, 10594. [Google Scholar] [CrossRef]
- Fleming, K.A.; Perrault, J.; Stacy, N.; Coppenrath, C.M.; Gainsbury, A.M. Heat, health and hatchlings: Associations of in situ nest temperatures with morphological and physiological characteristics of loggerhead sea turtle hatchlings from Florida. Conserv. Physiol. 2020, 8, coaa046. [Google Scholar] [CrossRef]
- Lu, H.; Jin, J.; Fan, H.; Dang, W. The magnitude of incubation temperature fluctuation affects the immunity of Chinese soft-shelled turtle (Pelodiscus sinensis) hatchlings. Aquac. Res. 2021. [Google Scholar] [CrossRef]
- Fisher, L.R.; Godfrey, M.H.; Owens, D.W. Incubation Temperature Effects on Hatchling Performance in the Loggerhead Sea Turtle (Caretta caretta). PLoS ONE 2014, 9, e114880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janes, D.E.; Organ, C.L.; Fujita, M.K.; Shedlock, A.M.; Edwards, S.V. Genome Evolution in Reptilia, the Sister Group of Mammals. Annu. Rev. Genom. Hum. Genet. 2010, 11, 239–264. [Google Scholar] [CrossRef] [Green Version]
- Radhakrishnan, S.; Literman, R.; Neuwald, J.; Severin, A.; Valenzuela, N. Transcriptomic responses to environmental temperature by turtles with temperature-dependent and genotypic sex determination assessed by RNAseq inform the genetic architecture of embryonic gonadal development. PLoS ONE 2017, 12, e0172044. [Google Scholar] [CrossRef]
- Deveson, I.W.; Holleley, C.E.; Blackburn, J.; Graves JA, M.; Mattick, J.S.; Waters, P.D.; Georges, A. Differential intron retention in Jumonji chromatin modifier genes is implicated in reptile temperature-dependent sex determination. Sci. Adv. 2017, 3, e1700731. [Google Scholar] [CrossRef] [Green Version]
- Yatsu, R.; Miyagawa, S.; Kohno, S.; Parrott, B.B.; Yamaguchi, K.; Ogino, Y.; Miyakawa, H.; Lowers, R.H.; Shigenobu, S.; Guillette, L.J., Jr.; et al. RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation. BMC Genom. 2016, 17, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentley, B.P.; Haas, B.J.; Tedeschi, J.; Berry, O. Loggerhead sea turtle embryos (Caretta caretta) regulate expression of stress response and developmental genes when exposed to a biologically realistic heat stress. Mol. Ecol. 2017, 26, 2978–2992. [Google Scholar] [CrossRef] [PubMed]
- Tezak, B.; Sifuentes-Romero, I.; Milton, S.; Wyneken, J. Identifying Sex of Neonate Turtles with Temperature-dependent Sex Determination via Small Blood Samples. Sci. Rep. 2020, 10, 1–8. [Google Scholar]
- Chow, J.C.; Anderson, P.E.; Shedlock, A.M. Sea Turtle Population Genomic Discovery: Global and Locus-Specific Signatures of Polymorphism, Selection, and Adaptive Potential. Genome Biol. Evol. 2019, 11, 2797–2806. [Google Scholar] [CrossRef] [Green Version]
- Godfrey, H.M.; Mrosovsky, N. Pivotal temperature for green sea turtles, Chelonia mydas, nesting in Suriname. Herpetol. J. 2006, 16, 55–61. [Google Scholar]
- Yntema, C.L.; Mrosovsky, N. Sexual Differentiation in Hatchling Loggerheads (Caretta caretta) Incubated at Different Controlled Temperatures. Herpetologica 1980, 36, 33–36. [Google Scholar]
- Harms, C.A.; McClellan-Green, P.; Godfrey, M.H.; Christiansen, E.F.; Broadhurst, H.J.; Godard-Codding, C.A.J. Crude Oil and Dispersant Cause Acute Clinicopathological Abnormalities in Hatchling Loggerhead Sea Turtles (Caretta caretta). Front. Vet. Sci. 2019, 6, 344. [Google Scholar] [CrossRef] [Green Version]
- Bembenek-Bailey, S.A.; Niemuth, J.N.; McClellan-Green, P.D.; Godfrey, M.H.; Harms, C.A.; Gracz, H.; Stoskopf, M.K. NMR Metabolomic Analysis of Skeletal Muscle, Heart, and Liver of Hatchling Loggerhead Sea Turtles (Caretta caretta) Experimentally Exposed to Crude Oil and/or Corexit. Metabolites 2019, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Bembenek Bailey, S.A.; Niemuth, J.N.; McClellan-Green, P.D.; Godfrey, M.H.; Harms, C.A.; Stoskopf, M.K. 1H-NMR metabolomic study of whole blood from hatchling loggerhead sea turtles (Caretta caretta) exposed to crude oil and/or Corexit. R. Soc. Open Sci. 2017, 4, 171433. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High -throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Smuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Shaffer, H.B.; Minx, P.; Warren, D.E.; Shedlock, A.M.; Thomson, R.C.; Valenzuela, N.; Abramyan, J.; Amemiya, C.T.; Badenhorst, D.; Biggar, K.K.; et al. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013, 14, R28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Pascual-Anaya, J.; Zadissa, A.; Li, W.; Niimura, Y.; Huang, Z.; Li, C.; White, S.; Xiong, Z.; Fang, D.; et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Genet. 2013, 45, 701–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.X.; Teng, J.; Zhao, Y.; Li, N.; Wang, H.; Ji, X.S. Gonad Transcriptome Analysis of High-Temperature-Treated Females and High-Temperature-Induced Sex-Reversed Neomales in Nile Tilapia. Int. J. Mol. Sci. 2018, 19, 689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, G.; Li, S.; Zhao, Q.; Chu, J.; Zhou, B.; Fan, S.; Shi, F.; Wei, X.; Hu, X.; Zheng, X.; et al. Transcriptomic and metabolomic insights into the variety of sperm storage in oviduct of egg layers. Poult. Sci. 2021, 100, 101087. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Buemio, A.; Chu, R.; Vafaee, M.; Crews, D. Epigenetic Control of Gonadal Aromatase (cyp19a1) in Temperature-Dependent Sex Determination of Red-Eared Slider Turtles. PLoS ONE 2013, 8, e63599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhen, T.; Metzger, K.; Schroeder, A.; Woodward, R. Expression of Putative Sex-Determining Genes during the Thermosensitive Period of Gonad Development in the Snapping Turtle, Chelydra serpentina. Sex. Dev. 2007, 1, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, C.; Ramsey, M.; Queen, J.; Crews, D. Expression ofSox9,Mis, andDmrt1 in the gonad of a species with temperature-dependent sex determination. Dev. Dyn. 2007, 236, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Hernández, V.; Marmolejo-Valencia, A.; Merchant-Larios, H. Exogenous estradiol alters gonadal growth and timing ofmtemperature sex determination in gonads of sea turtle. Dev. Biol. 2015, 408, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, A.L.; Metzger, K.J.; Miller, A.; Rhen, T. A Novel Candidate Gene for Temperature-Dependent Sex Determination in the Common Snapping Turtle. Genetics 2016, 203, 557–571. [Google Scholar] [CrossRef] [Green Version]
- Mohammed, H.; D’Santos, C.; Serandour, A.A.; Ali, R.; Brown, G.D.; Atkins, A.; Palacio, O.R.; Holmes, K.-A.; Theodorou, V.; Robinson, J.L.; et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2021, 3, 342–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luangpraseuth-Prosper, A.; Lesueur, E.; Jouneau, L.; Pailhoux, E.; Cotinot, C.; Mandon-Pépin, B. TOPAZ1, a germ cell specific factor, is essential for male meiotic progression. Dev. Biol. 2015, 406, 158–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arango, N.A.; Li, L.; Dabir, D.; Nicolau, F.; Pieretti-Vanmarcke, R.; Koehler, C.; McCarrey, J.R.; Lu, N.; Donahoe, P.K. Meiosis I arrest abnormalities lead to severe oligozoospermia in meiosis 1 arresting protein (M1ap)-deficient mice. Biol. Reprod. 2013, 88, 76. [Google Scholar] [CrossRef]
- Ge, C.; Ye, J.; Weber, C.; Sun, W.; Zhang, H.; Zhou, Y.; Cai, C.; Qian, G.; Capel, B. The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science 2018, 360, 645–648. [Google Scholar] [CrossRef] [Green Version]
- Weber, C.; Zhou, Y.; Lee, J.G.; Looger, L.L.; Qian, G.; Ge, C.; Capel, B. Temperature-dependent sex determination is mediated by pSTAT3 repression of Kdm6b. Science 2020, 368, 303–306. [Google Scholar] [CrossRef]
- Purnell, B.A. How egg temperature sets sex. Science 2020, 368, 278. [Google Scholar]
- Boyle, M.C.; Fitzsimmons, N.N.; Limpus, C.J.; Kelez, S.; Velez-Zuazo, X.; Waycott, M. Evidence for transoceanic migrations by loggerhead sea turtles in the southern Pacific Ocean. Proc. R. Soc. B Biol. Sci. 2009, 276, 1993–1999. [Google Scholar] [CrossRef] [Green Version]
- Bowen, B.W.; Kamezaki, N.; Limpus, C.J.; Hughes, G.R.; Meylan, A.B.; Avise, J.C. Global Phylogeography of the Loggerhead Turtle (Caretta caretta) as Indicated by Mitochondrial DNA Haplotypes. Evolution 1994, 48, 1820. [Google Scholar]
- Lohmann, K.J.; Putman, N.F.; Lohmann, C.M.F. Geomagnetic imprinting: A unifying hypothesis of long-distance natal homing in salmon and sea turtles. Proc. Natl. Acad. Sci. USA 2008, 105, 19096–19101. [Google Scholar] [CrossRef] [Green Version]
- Shamblin, B.M.; Bolten, A.; Abreu-Grobois, F.A.; Bjorndal, K.; Cardona, L.; Carreras, C.; Clusa, M.; Monzón-Argüello, C.; Nairn, C.J.; Nielsen, J.T.; et al. Geographic Patterns of Genetic Variation in a Broadly Distributed Marine Vertebrate: New Insights into Loggerhead Turtle Stock Structure from Expanded Mitochondrial DNA Sequences. PLoS ONE 2014, 9, e85956. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.A.; Lau, J.C.S.; Hogben, H.J.; Biskup, T.; Kattnig, D.R.; Hore, P.J. Alternative radical pairs for cryptochrome-based magnetoreception. J. R. Soc. Interface 2014, 11, 20131063. [Google Scholar] [CrossRef] [Green Version]
- Boss, J.; Liedvogel, M.; Lundberg, M.; Olsson, P.; Reischke, N.; Naurin, S.; Åkesson, S.; Hasselquist, D.; Wright, A.; Grahn, M.; et al. Gene expression in the brain of a migratory songbird during breeding and migration. Mov. Ecol. 2016, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Mueller, J.C.; Pulido, F.; Kempenaers, B. Identification of a gene associated with avian migratory behaviour. Proc. R. Soc. B Biol. Sci. 2011, 278, 2848–2856. [Google Scholar] [CrossRef] [PubMed]
- Toews, D.P.L.; Taylor, S.A.; Streby, H.M.; Kramer, G.R.; Lovette, I.J. Selection on VPS13A linked to migration in a songbird. Proc. Natl. Acad. Sci. USA 2019, 116, 18272–18274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouw, L.G.; Reading, N.S.; Jenson, S.D.; Lim, M.S.; Elenitoba-Johnson, K.S.J. Expression of the Rho-family GTPase gene RHOF in lymphocyte subsets and malignant lymphomas. Br. J. Haematol. 2005, 129, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Tokue, M.; Ikami, K.; Mizuno, S.; Takagi, C.; Miyagi, A.; Takada, R.; Noda, C.; Kitadate, Y.; Hara, K.; Mizuguchi, H.; et al. SHISA6 Confers Resistance to Differentiation-Promoting Wnt/β-Catenin Signaling in Mouse Spermatogenic Stem Cells. Stem Cell Rep. 2017, 8, 561–575. [Google Scholar] [CrossRef] [Green Version]
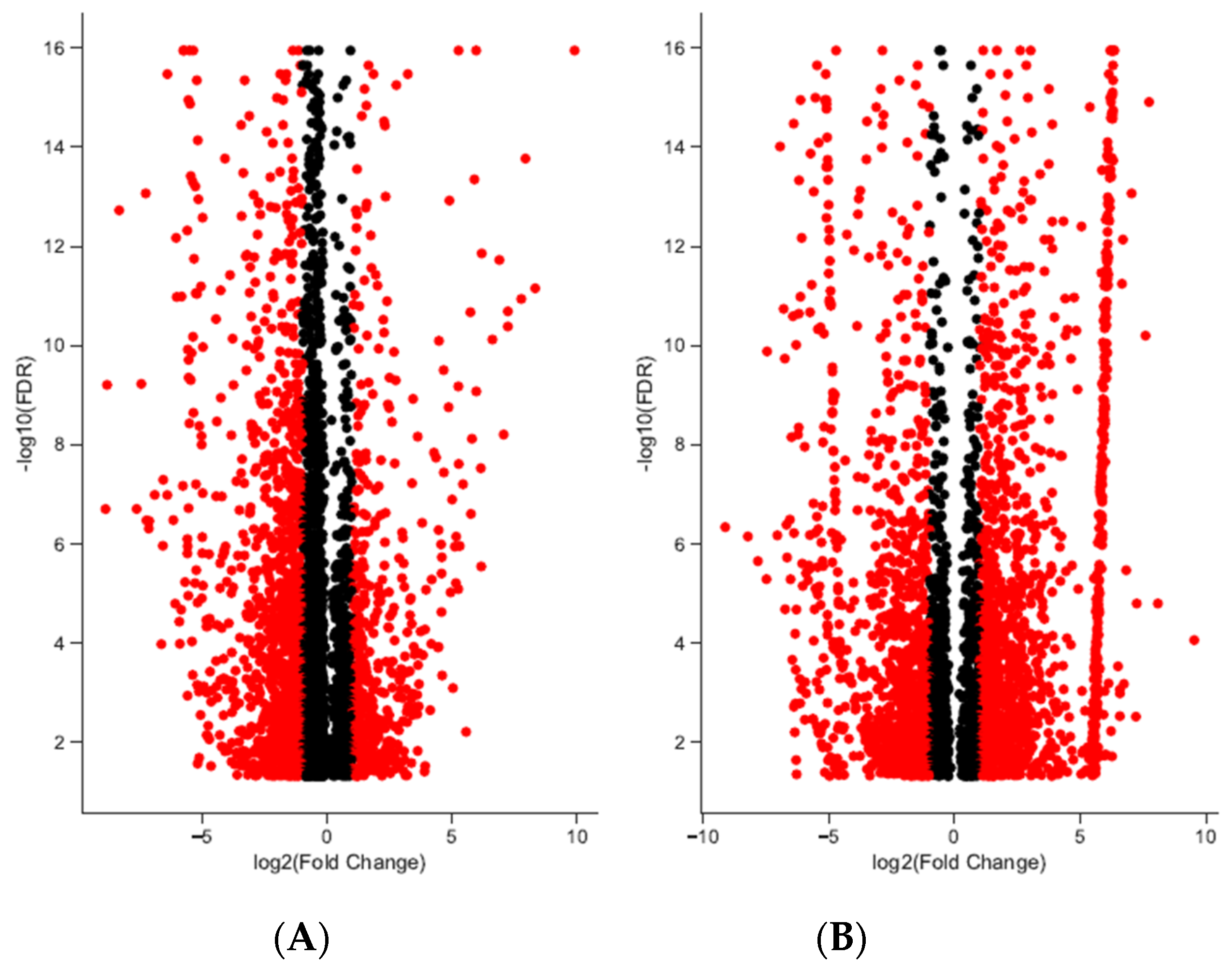
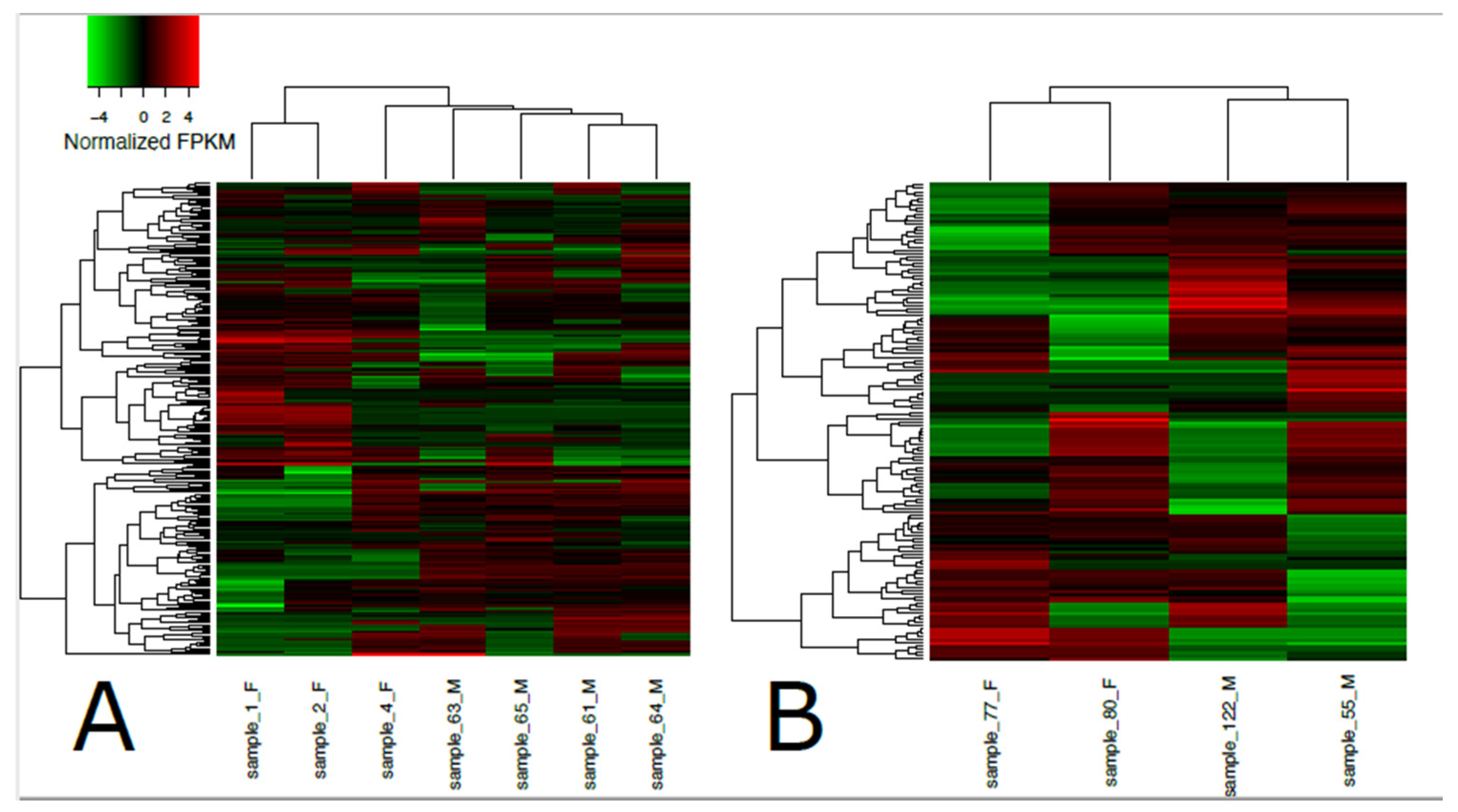

| Sample ID | Sex | Raw Reads | Reads Mapped to Transcriptome (%) |
|---|---|---|---|
| Brain | |||
| 1 | Female | 123,316,446 | 91.57% |
| 2 | Female | 73,620,245 | 91.48% |
| 4 | Female | 65,569,443 | 91.44% |
| 61 | Male | 102,155,296 | 91.22% |
| 63 | Male | 17,845,489 | 90.39% |
| 64 | Male | 96,913,331 | 91.43% |
| 65 | Male | 102,423,395 | 91.20% |
| Gonad | |||
| 77 | Female | 51,399,125 | 82.33% |
| 80 | Female | 50,746,307 | 83.58% |
| 55 | Male | 46,557,205 | 83.88% |
| 122 | Male | 11,018,312 | 84.06% |
| Brain | ||||
| Accession | Gene | Description | q-value | Fold change |
| XP_007072694 | SEPT1 | septin 1 | 0 | 727.96 |
| XP_008170752 | LOC101945970 | shugoshin-like 2 | 1.75 × 10−2 | 3.17 |
| XP_007061912 | MAPKAPK3 | MAP kinase- activated kinase 3 | 9.67 × 10−3 | 5.58 |
| XP_008177295 | ESRRG | estrogen-related receptor gamma isoform X3 | 2.23 × 10−2 | 6.53 |
| XP_007054941 | JARID2 | Jumonji isoform X1 | 4.49 × 10−11 | 2.09 |
| XP_007060139 | TOPAZ1 | testis- and ovary- specific PAZ domain-containing 1 | 2.41 × 10−5 | 23.65 |
| XP_007063347 | GREB1 | GREB1 isoform X1 | 4.01 × 10−2 | 14.79 |
| XP_007063772 | KDM3A | lysine-specific demethylase 3A | 0 | 61.13 |
| XP_007055513 | RPS6KA5 | ribosomal S6 kinase alpha-5 isoform X1 | 2.90 × 10−3 | 10.60 |
| XP_008174364 | KDM6B | lysine-specific demethylase 6B | 5.60 × 10−10 | 3.13 |
| XP_007056370 | HSD17B7 | 3-keto-steroid reductase | 3.77 × 10−2 | 5.31 |
| XP_007069240 | MSMP | prostate-associated microsemino | 2.83 × 10−4 | 12.36 |
| Gonad | ||||
| Accession | Gene | Description | q-value | Fold change |
| XP_007060743 | RSPO1 | R-spondin-1 isoform X2 | 4.63 × 10−3 | 5.51 |
| XP_005283194 | CBX2 | Chromobox homolog 2 | 1.20 × 10−2 | 2.39 |
| XP_005282573 | FOXL2 | forkhead box L2 | 0 | 64.96 |
| XP_008170093 | GATA4 | transcription factor GATA-4 isoform X2 | 3.09 × 10−2 | 8.95 |
| XP_007065874 | AMHR2 | anti-Muellerian hormone type-2 receptor | 1.22 × 10−6 | 5.59 |
| XP_006124717 | LHX9 | LIM homeobox Lhx9 isoform X4 | 1.40 × 10−11 | 3.88 |
| XP_007063819 | BCR | breakpoint cluster region isoform X1 | 7.92 × 10−4 | 2.18 |
| XP_007065642 | DMRT1 | doublesex and mab-3 related transcription factor 1 | 1.67 × 10−3 | 70.35 |
| Biological Process Term | GO:ID | Brain q-Value | Gonad q-Value |
|---|---|---|---|
| Development of primary sexual characteristics | 0045137 | 2.5527 × 10−4 | 2.2747 × 10−2 |
| Embryo development ending in birth or egg hatching | 0009792 | 4.8274 × 10−7 | 3.7780 × 10−3 |
| Female gamete generation | 0007292 | 2.2815 × 10−2 | |
| Female sex differentiation | 0046660 | 1.1049 × 10−2 | |
| Germ cell development | 0007281 | 5.4465 × 10−5 | 2.7750 × 10−3 |
| Gonad development | 0008406 | 2.5527 × 10−4 | 2.2747 × 10−2 |
| Male gamete generation | 0048232 | 1.1888 × 10−4 | 1.1216 × 10−3 |
| Male sex differentiation | 0046661 | 4.6000 × 10−2 | |
| response to steroid hormone | 0048545 | 4.4088 × 10−8 | 8.4302 × 10−4 |
| Sex differentiation | 0007548 | 1.1821 × 10−4 | 2.7750 × 10−3 |
| Steroid biosynthetic process | 0006694 | 1.1049 × 10−2 | 2.2747 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chow, J.C.; Kyritsis, N.; Mills, M.; Godfrey, M.H.; Harms, C.A.; Anderson, P.E.; Shedlock, A.M. Tissue and Temperature-Specific RNA-Seq Analysis Reveals Genomic Versatility and Adaptive Potential in Wild Sea Turtle Hatchlings (Caretta caretta). Animals 2021, 11, 3013. https://doi.org/10.3390/ani11113013
Chow JC, Kyritsis N, Mills M, Godfrey MH, Harms CA, Anderson PE, Shedlock AM. Tissue and Temperature-Specific RNA-Seq Analysis Reveals Genomic Versatility and Adaptive Potential in Wild Sea Turtle Hatchlings (Caretta caretta). Animals. 2021; 11(11):3013. https://doi.org/10.3390/ani11113013
Chicago/Turabian StyleChow, Julie C., Nia Kyritsis, Micah Mills, Matthew H. Godfrey, Craig A. Harms, Paul E. Anderson, and Andrew M. Shedlock. 2021. "Tissue and Temperature-Specific RNA-Seq Analysis Reveals Genomic Versatility and Adaptive Potential in Wild Sea Turtle Hatchlings (Caretta caretta)" Animals 11, no. 11: 3013. https://doi.org/10.3390/ani11113013
APA StyleChow, J. C., Kyritsis, N., Mills, M., Godfrey, M. H., Harms, C. A., Anderson, P. E., & Shedlock, A. M. (2021). Tissue and Temperature-Specific RNA-Seq Analysis Reveals Genomic Versatility and Adaptive Potential in Wild Sea Turtle Hatchlings (Caretta caretta). Animals, 11(11), 3013. https://doi.org/10.3390/ani11113013






