Detoxification, Hydrogen Sulphide Metabolism and Wound Healing Are the Main Functions That Differentiate Caecum Protein Expression from Ileum of Week-Old Chicken
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of Chicken Samples
2.2. Protein and RNA Purification from Chicken Caecal and Ileal Tissue
2.3. Protein Mass Spectrometry
2.4. Quantitative Reverse-Transcribed PCR (qRT-PCR)
2.5. Statistics
2.6. Ethics Approval and Consent to Participate
3. Results
3.1. Gross Comparison of Protein Expression in the Ileum and Caecum
3.2. Ileum-Specific Proteins
3.3. Caecum-Specific Proteins
3.4. Functional Analysis of Caecum-Specific Proteins
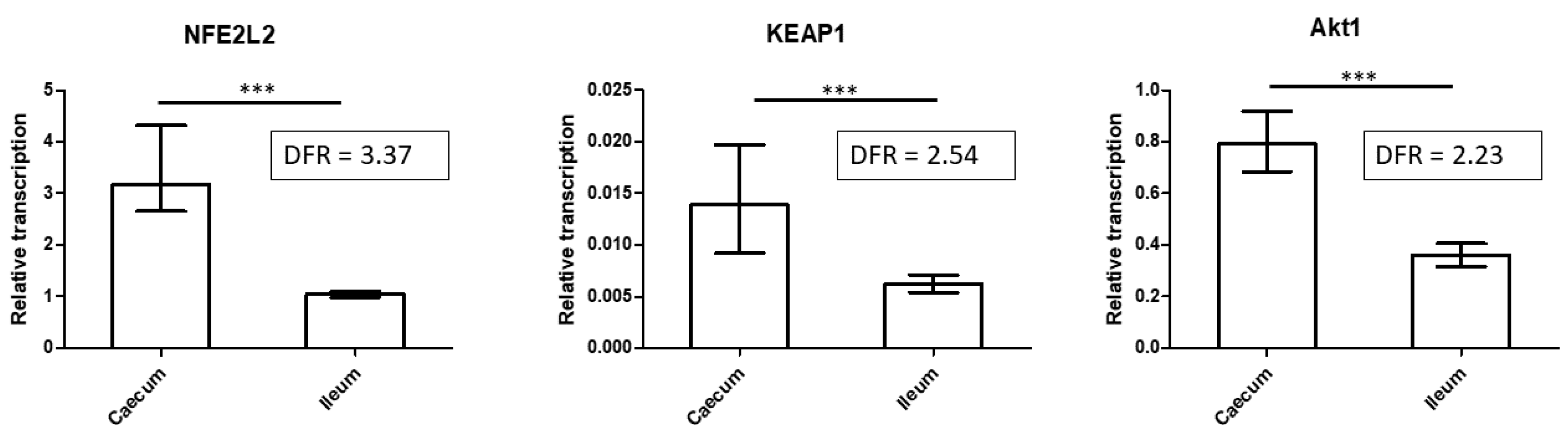
3.5. Expression of Key Regulatory Proteins of Oxidative Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Svihus, B.; Choct, M.; Classen, H. Function and nutritional roles of the avian caeca: A review. World’s Poult. Sci. J. 2013, 69, 249–264. [Google Scholar] [CrossRef]
- Isshiki, Y.; Nakahiro, Y. Effect of Ceca Removal on Water Absorption in Chickens. Jpn. Poult. Sci. 1975, 12, 271–273. [Google Scholar] [CrossRef] [Green Version]
- Sanders, K.M.; Koh, S.D.; Ro, S.; Ward, S.M. Regulation of gastrointestinal motility—insights from smooth muscle biology. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 633–645. [Google Scholar] [CrossRef] [Green Version]
- Clench, M.H. The Avian Cecum: Update and Motility Review. J. Exp. Zool. 1999, 283, 441–447. [Google Scholar] [CrossRef]
- Videnska, P.; Faldynova, M.; Juricova, H.; Babak, V.; Sisak, F.; Havlickova, H.; Rychlik, I. Chicken faecal microbiota and disturbances induced by single or repeated therapy with tetracycline and streptomycin. BMC Vet. Res. 2013, 9, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Videnska, P.; Sedlar, K.; Lukac, M.; Faldynova, M.; Gerzova, L.; Cejkova, D.; Sisak, F.; Rychlik, I. Succession and Replacement of Bacterial Populations in the Caecum of Egg Laying Hens over Their Whole Life. PLoS ONE 2014, 9, e115142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pol, A.; Renkema, G.H.; Tangerman, A.; Winkel, E.G.; Engelke, U.F.; De Brouwer, A.P.M.; Lloyd, K.C.; Araiza, R.S.; Heuvel, L.V.D.; Omran, H.; et al. Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Nat. Genet. 2018, 50, 120–129. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Kelly, C.; Colgan, S.P. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am. J. Physiol.-Cell Physiol. 2015, 309, C350–C360. [Google Scholar] [CrossRef] [Green Version]
- Colgan, S.P.; Taylor, C. Hypoxia: An alarm signal during intestinal inflammation. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 281–287. [Google Scholar] [CrossRef] [Green Version]
- Volf, J.; Polansky, O.; Sekelova, Z.; Velge, P.; Schouler, C.; Kaspers, B.; Rychlik, I. Gene expression in the chicken caecum is dependent on microbiota composition. Veter. Res. 2017, 48, 85. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Volf, J.; Polansky, O.; Varmuzova, K.; Gerzova, L.; Sekelova, Z.; Faldynova, M.; Babak, V.; Medvecky, M.; Smith, A.L.; Kaspers, B.; et al. Transient and Prolonged Response of Chicken Cecum Mucosa to Colonization with Different Gut Microbiota. PLoS ONE 2016, 11, e0163932. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Bertocchi, M.; Sirri, F.; Palumbo, O.; Luise, D.; Maiorano, G.; Bosi, P.; Trevisi, P. Exploring Differential Transcriptome between Jejunal and Cecal Tissue of Broiler Chickens. Animals 2019, 9, 221. [Google Scholar] [CrossRef] [Green Version]
- Tyska, M.J.; Mackey, A.T.; Huang, J.-D.; Copeland, N.G.; Jenkins, N.A.; Mooseker, M.S. Myosin-1a Is Critical for Normal Brush Border Structure and Composition. Mol. Biol. Cell 2005, 16, 2443–2457. [Google Scholar] [CrossRef] [Green Version]
- Sim, L.; Quezada-Calvillo, R.; Sterchi, E.E.; Nichols, B.L.; Rose, D.R. Human Intestinal Maltase–Glucoamylase: Crystal Structure of the N-Terminal Catalytic Subunit and Basis of Inhibition and Substrate Specificity. J. Mol. Biol. 2008, 375, 782–792. [Google Scholar] [CrossRef]
- Danielsen, E.M. Tyrosine Sulfation, a Post-Translational Modification of Microvillar Enzymes in the Small Intestinal Entero-cyte. EMBO J. 1987, 6, 2891–2896. [Google Scholar] [CrossRef] [PubMed]
- Sim, L.; Willemsma, C.; Mohan, S.; Naim, H.Y.; Pinto, B.M.; Rose, D.R. Structural Basis for Substrate Selectivity in Human Maltase-Glucoamylase and Sucrase-Isomaltase N-terminal Domains. J. Biol. Chem. 2010, 285, 17763–17770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Praslickova, D.; Torchia, E.C.; Sugiyama, M.G.; Magrane, E.J.; Zwicker, B.L.; Kolodzieyski, L.; Agellon, L.B. The Ileal Lipid Binding Protein Is Required for Efficient Absorption and Transport of Bile Acids in the Distal Portion of the Murine Small Intestine. PLoS ONE 2012, 7, e50810. [Google Scholar] [CrossRef] [Green Version]
- Huang, E.; Hynes, M.J.; Zhang, T.; Ginestier, C.; Dontu, G.; Appelman, H.; Fields, J.Z.; Wicha, M.S.; Boman, B.M. Aldehyde Dehydrogenase 1 Is a Marker for Normal and Malignant Human Colonic Stem Cells (SC) and Tracks SC Overpopulation during Colon Tumorigenesis. Cancer Res. 2009, 69, 3382–3389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vassalli, G. Aldehyde Dehydrogenases: Not Just Markers, but Functional Regulators of Stem Cells. Stem Cells Int. 2019, 2019, 3904645. [Google Scholar] [CrossRef] [Green Version]
- Taskoparan, B.; Seza, E.G.; Demirkol, S.; Tunçer, S.; Stefek, M.; Gure, A.O.; Banerjee, S. Opposing roles of the aldo-keto reductases AKR1B1 and AKR1B10 in colorectal cancer. Cell. Oncol. (Dordr.) 2017, 40, 563–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanson, J.D.; Forwood, J.K. Structural Characterisation of FabG from Yersinia pestis, a Key Component of Bacterial Fatty Acid Synthesis. PLoS ONE 2015, 10, e0141543. [Google Scholar] [CrossRef] [PubMed]
- Mayoral, J.G.; Leonard, K.T.; DeFelipe, L.A.; Turjansksi, A.G.; Nouzova, M.; Noriegal, F.G. Functional analysis of a mosquito short-chain dehydrogenase cluster. Arch. Insect Biochem. Physiol. 2013, 82, 96–115. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Li, Y.; Yang, L.; Zhang, D.; Liu, Z.; Wang, Y.; Han, R.; Li, G.; Li, Z.; Tian, Y.; et al. Identification of a Novel Lipid Metabolism-Associated Hepatic Gene Family Induced by Estrogen via ERα in Chicken (Gallus gallus). Front. Genet. 2020, 11, 271. [Google Scholar] [CrossRef]
- Yodoi, J.; Matsuo, Y.; Tian, H.; Masutani, H.; Inamoto, T. Anti-Inflammatory Thioredoxin Family Proteins for Medicare, Healthcare and Aging Care. Nutrients 2017, 9, 1081. [Google Scholar] [CrossRef] [Green Version]
- Bindoli, A.; Fukuto, J.M.; Forman, H.J. Thiol Chemistry in Peroxidase Catalysis and Redox Signaling. Antioxid. Redox Signal. 2008, 10, 1549–1564. [Google Scholar] [CrossRef]
- Maclean, K.N.; Greiner, L.S.; Evans, J.R.; Sood, S.K.; Lhotak, S.; Markham, N.E.; Stabler, S.P.; Allen, R.H.; Austin, R.C.; Balasubramaniam, V.; et al. Cystathionine Protects against Endoplasmic Reticulum Stress-induced Lipid Accumulation, Tissue Injury, and Apoptotic Cell Death. J. Biol. Chem. 2012, 287, 31994–32005. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, Y.; Zhang, L.; Zhang, D.; Bai, L.; Kong, W.; Huang, Y.; Tang, C.; Du, J.; Jin, H. L-Cystathionine Protects against Homocysteine-Induced Mitochondria-Dependent Apoptosis of Vascular Endothelial Cells. Oxidative Med. Cell. Longev. 2019, 2019, 1253289. [Google Scholar] [CrossRef]
- Gobbi, R.P.; De Francesco, N.; Bondar, C.; Muglia, C.; Chirdo, F.; Rumbo, M.; Rocca, A.; Toscano, M.; Sambuelli, A.; A Rabinovich, G.; et al. A galectin-specific signature in the gut delineates Crohn’s disease and ulcerative colitis from other human inflammatory intestinal disorders. BioFactors 2016, 42, 93–105. [Google Scholar] [CrossRef]
- Thijssen, V.; Postel, R.; Brandwijk, R.J.M.G.E.; Dings, R.; Nesmelova, I.; Satijn, S.; Verhofstad, N.; Nakabeppu, Y.; Baum, L.G.; Bakkers, J.; et al. Galectin-1 is essential in tumor angiogenesis and is a target for antiangiogenesis therapy. Proc. Natl. Acad. Sci. USA 2006, 103, 15975–15980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Liu, M.; Li, Z.; Wu, X.-B.; Wang, Y.; Wang, Y.; Nie, M.; Huang, F.; Ju, J.; Ma, C.; et al. LYAR promotes colorectal cancer cell mobility by activating galectin-1 expression. Oncotarget 2015, 6, 32890–32901. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Ye, Y.; Jia, J.; He, Y.; Yang, Z.; Zhu, X.; Huang, H.; Wang, W.; Geng, L.; Yin, S.; et al. Galectin-1-induced tolerogenic dendritic cells combined with apoptotic lymphocytes prolong liver allograft survival. Int. Immunopharmacol. 2018, 65, 470–482. [Google Scholar] [CrossRef]
- Garin, M.I.; Chu, C.-C.; Golshayan, D.; Cernuda-Morollón, E.; Wait, R.; Lechler, R.I. Galectin-1: A key effector of regulation mediated by CD4+CD25+ T cells. Blood 2007, 109, 2058–2065. [Google Scholar] [CrossRef] [Green Version]
- Fox, J.W.; Mayer, U.; Nischt, R.; Aumailley, M.; Reinhardt, D.; Wiedemann, H.; Mann, K.; Timpl, R.; Krieg, T.; Engel, J. Re-combinant Nidogen Consists of Three Globular Domains and Mediates Binding of Laminin to Collagen Type IV. EMBO J. 1991, 10, 3137–3146. [Google Scholar] [CrossRef]
- Reinhardt, D.; Mann, K.; Nischt, R.; Fox, J.; Chu, M.; Krieg, T.; Timpl, R. Mapping of nidogen binding sites for collagen type IV, heparan sulfate proteoglycan, and zinc. J. Biol. Chem. 1993, 268, 10881–10887. [Google Scholar] [CrossRef]
- Pöschl, E.; Fox, J.; Block, D.; Mayer, U.; Timpl, R. Two non-contiguous regions contribute to nidogen binding to a single EGF-like motif of the laminin gamma 1 chain. EMBO J. 1994, 13, 3741–3747. [Google Scholar] [CrossRef]
- Pankov, R.; Yamada, K. Fibronectin at a glance. J. Cell Sci. 2002, 115, 3861–3863. [Google Scholar] [CrossRef] [Green Version]
- Shankar, J.; Messenberg, A.; Chan, J.; Underhill, T.M.; Foster, L.J.; Nabi, I.R. Pseudopodial Actin Dynamics Control Epithelial-Mesenchymal Transition in Metastatic Cancer Cells. Cancer Res. 2010, 70, 3780–3790. [Google Scholar] [CrossRef] [Green Version]
- Mejillano, M.R.; Kojima, S.-I.; Applewhite, D.A.; Gertler, F.B.; Svitkina, T.M.; Borisy, G.G. Lamellipodial Versus Filopodial Mode of the Actin Nanomachinery: Pivotal Role of the Filament Barbed End. Cell 2004, 118, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Ammer, A.G.; Weed, S.A. Cortactin branches out: Roles in regulating protrusive actin dynamics. Cell Motil. Cytoskelet. 2008, 65, 687–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirfel, J.; Pantelis, D.; Kabba, M.; Kahl, P.; Röper, A.; Kalff, J.C.; Buettner, R. Impaired intestinal wound healing in Fhl2-deficient mice is due to disturbed collagen metabolism. Exp. Cell Res. 2008, 314, 3684–3691. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, B.; Guo, Z.; Sardet, C.; Zou, B.; Lam, C.S.; Li, J.; He, M.-L.; Lan, H.-Y.; Pang, R.; et al. Four-and-a-half LIM protein 2 promotes invasive potential and epithelial-mesenchymal transition in colon cancer. Carcinogenesis 2010, 31, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Wixler, V. The role of FHL2 in wound healing and inflammation. FASEB J. 2019, 33, 7799–7809. [Google Scholar] [CrossRef] [PubMed]
- Aktar, R.; Peiris, M.; Fikree, A.; Cibert-Goton, V.; Walmsley, M.; Tough, I.R.; Watanabe, P.; Araujo, E.J.D.A.; Mohammed, S.D.; Delalande, J.-M.; et al. The extracellular matrix glycoprotein tenascin-X regulates peripheral sensory and motor neurones. J. Physiol. 2018, 596, 4237–4251. [Google Scholar] [CrossRef]
- Kleist, B.; Poetsch, M. Neuroendocrine differentiation: The mysterious fellow of colorectal cancer. World J. Gastroenterol. 2015, 21, 11740–11747. [Google Scholar] [CrossRef]
- Gunawardene, A.; Corfe, B.M.; Staton, C.A. Classification and functions of enteroendocrine cells of the lower gastrointestinal tract. Int. J. Exp. Pathol. 2011, 92, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Attene-Ramos, M.S.; Wagner, E.D.; Gaskins, H.R.; Plewa, M.J. Hydrogen Sulfide Induces Direct Radical-Associated DNA Damage. Mol. Cancer Res. 2007, 5, 455–459. [Google Scholar] [CrossRef] [Green Version]
- Attene-Ramos, M.S.; Nava, G.M.; Muellner, M.G.; Wagner, E.D.; Plewa, M.J.; Gaskins, H.R. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ. Mol. Mutagen. 2010, 51, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Wallace, J.L.; Ferraz, J.G.; Muscara, M. Hydrogen Sulfide: An Endogenous Mediator of Resolution of Inflammation and Injury. Antioxid. Redox Signal. 2012, 17, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Flannigan, K.L.; Ferraz, J.G.P.; Wang, R.; Wallace, J.L. Enhanced Synthesis and Diminished Degradation of Hydrogen Sulfide in Experimental Colitis: A Site-Specific, Pro-Resolution Mechanism. PLoS ONE 2013, 8, e71962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirata, I.; Naito, Y.; Takagi, T.; Mizushima, K.; Suzuki, T.; Omatsu, T.; Handa, O.; Ichikawa, H.; Ueda, H.; Yoshikawa, T. Endogenous Hydrogen Sulfide Is an Anti-inflammatory Molecule in Dextran Sodium Sulfate-Induced Colitis in Mice. Dig. Dis. Sci. 2011, 56, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Motta, J.-P.; Flannigan, K.L.; Agbor, T.A.; Beatty, J.K.; Blackler, R.W.; Workentine, M.L.; Da Silva, G.J.; Wang, R.; Buret, A.G.; Wallace, J.L. Hydrogen Sulfide Protects from Colitis and Restores Intestinal Microbiota Biofilm and Mucus Production. Inflamm. Bowel Dis. 2015, 21, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Calvert, J.; Jha, S.; Gundewar, S.; Elrod, J.; Ramachandran, A.; Pattillo, C.B.; Kevil, C.; Lefer, D.J. Hydrogen Sulfide Mediates Cardioprotection Through Nrf2 Signaling. Circ. Res. 2009, 105, 365–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-Antioxidant Response Element Signaling Pathway and Its Activation by Oxidative Stress. J. Biol. Chem. 2009, 284, 13291–13295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Chen, Y.; Sternberg, P.; Cai, J. Essential Roles of the PI3 Kinase/Akt Pathway in Regulating Nrf2-Dependent Anti-oxidant Functions in the RPE. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1671–1678. [Google Scholar] [CrossRef] [Green Version]
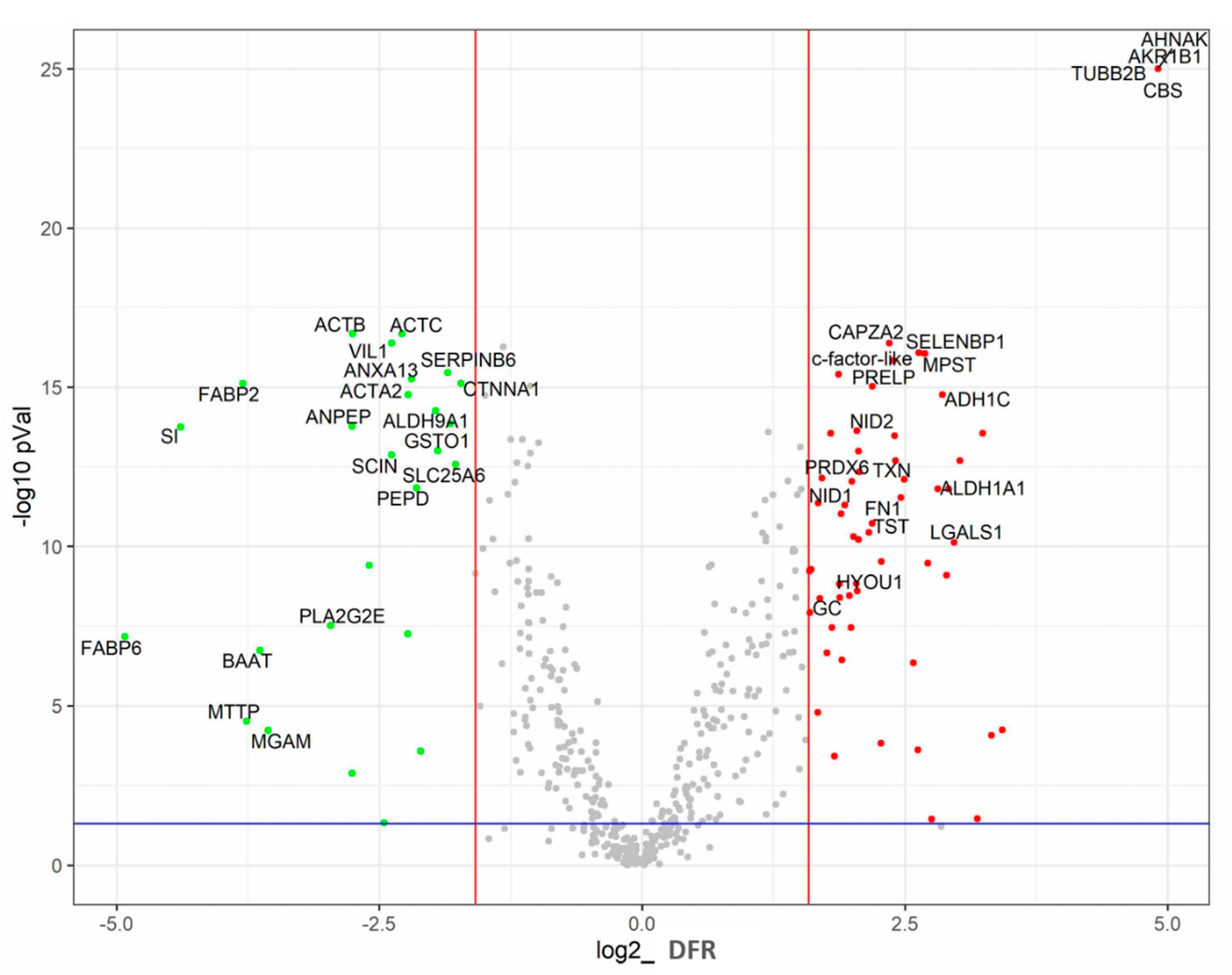

| Protein Annotation | Protein | p-Value | Abundance in Ileum [%] | DFR * | qRT-PCR Fold |
|---|---|---|---|---|---|
| ENSGALP00000010239 | ACTA2 | 1.73 × 10−15 | 4.94 | 4.67 | -# |
| ENSGALP00000039176 | ACTB | 2.09 × 10−17 | 4.82 | 6.75 | NS |
| ENSGALP00000018042 | ACTC | 2.09 × 10−17 | 4.64 | 4.87 | - |
| ENSGALP00000002196 | FABP6 | 6.95 × 10−8 | 4.12 | 311.34 | 2.43 |
| ENSGALP00000037398 | VIL1 | 4.18 × 10−17 | 0.60 | 5.21 | 6.43 |
| ENSGALP00000019528 | FABP2 | 7.73 × 10−16 | 0.50 | 13.89 | - |
| ENSGALP00000015467 | SI | 1.78 × 10−14 | 0.40 | 20.94 | 313.41 |
| ENSGALP00000003847 | CTNNA1 | 7.72 × 10−16 | 0.27 | 3.30 | NS |
| ENSGALP00000037173 | SCIN | 1.33 × 10−13 | 0.26 | 5.21 | - |
| ENSGALP00000005520 | ALDH9A1 | 5.55 × 10−15 | 0.2 | 3.90 | - |
| ENSGALP00000020953 | SERPINB6 | 3.45 × 10−16 | 0.17 | 3.61 | - |
| ENSGALP00000000256 | MGAM | 5.94 × 10−5 | 0.16 | 11.77 | - |
| ENSGALP00000007819 | PEPD | 1.48 × 10−12 | 0.13 | 4.42 | - |
| ENSGALP00000032098 | BAAT | 1.85 × 10−7 | 0.12 | 12.42 | 7.00 |
| ENSGALP00000035815 | SLC25A6 | 2.67 × 10−13 | 0.12 | 3.42 | - |
| ENSGALP00000008315 | ANPEP | 1.68 × 10−14 | 0.10 | 6.76 | - |
| ENSGALP00000013682 | GSTO1 | 9.86 × 10−14 | 0.10 | 3.85 | - |
| ENSGALP00000022884 | PLA2G2E | 3.05 × 10−8 | 0.09 | 7.81 | - |
| ENSGALP00000026341 | ANXA13 | 5.55 × 10−16 | 0.09 | 4.58 | - |
| ENSGALP00000036834 | MTTP | 3.05 × 10−5 | 0.09 | 13.57 | - |
| Group Description | No. of Proteins | % Expression out of Ileum-Specific Proteins | Proteins |
|---|---|---|---|
| Actin and actin-binding | 7 | 70.13 | ACTA2; ACTB; ACTC; VIL1; CTNNA1; SCIN; MYO1A |
| Bile metabolism and transport | 2 | 19.04 | FABP6; BAAT |
| Digestion enzymes | 4 | 3.53 | SI; MGAM; PEPD; ANPEP |
| Lipid metabolism and transport | 4 | 3.51 | FABP2; PLA2G2E; MTTP; PLIN4 |
| Other | 8 | 3.80 | SERPINB6; ANXA13; GSTO1; SLC25A6; RPS8; RPS8; GDA; SORD |
| Protein Annotation | Protein | p-Value | Abundance in Caecum [%] | DFR * | qRT-PCR Fold | Influenced by Microbiota Colonisation # |
|---|---|---|---|---|---|---|
| ENSGALP00000020275 | GAL-1 | 7.49 × 10−11 | 0.63 | 7.83 | 2.40 | |
| ENSGALP00000025280 | TXN | 7.89 × 10−13 | 0.59 | 5.62 | 6.39 | UP |
| ENSGALP00000001060 | c-factor-like | 8.37 × 10−17 | 0.48 | 6.20 | - | |
| ENSGALP00000026062 | CBS | NA | 0.43 | 500.00 | 3488.35 | UP |
| ENSGALP00000001274 | SELENBP1 | 8.72 × 10−17 | 0.36 | 6.44 | 14.72 | |
| ENSGALP00000019979 | ADH1C | 1.73 × 10−15 | 0.21 | 7.24 | 31.09 | DOWN |
| ENSGALP00000036423 | TUBB2B | NA | 0.20 | 500.00 | - | |
| ENSGALP00000004816 | PRDX6 | 7.23 × 10−13 | 0.20 | 3.28 | 13.14 | |
| ENSGALP00000029947 | TST | 3.65 × 10−11 | 0.19 | 4.47 | 4.90 | UP |
| ENSGALP00000017869 | NID1 | 5.04 × 10−12 | 0.18 | 3.81 | - | |
| ENSGALP00000005480 | AKR1B1L | NA | 0.18 | 500.00 | - | DOWN |
| ENSGALP00000005607 | PRELP | 9.41 × 10−16 | 0.18 | 4.56 | - | |
| ENSGALP00000024396 | ALDH1A1 | 1.58 × 10−12 | 0.17 | 7.02 | 16.55 | DOWN |
| ENSGALP00000005654 | FN1 | 2.98 × 10−12 | 0.16 | 5.50 | 2.37 | |
| ENSGALP00000020214 | NID2 | 2.37 × 10−14 | 0.14 | 4.12 | - | |
| ENSGALP00000020373 | MPST | 8.72 × 10−17 | 0.14 | 6.46 | 7.43 | |
| ENSGALP00000023289 | AHNAK | NA | 0.12 | 500.00 | 2.19 | |
| ENSGALP00000039757 | HYOU1 | 2.52 × 10−9 | 0.12 | 4.13 | - | |
| ENSGALP00000015277 | CAPZA2 | 4.18 × 10−17 | 0.11 | 5.11 | - | |
| ENSGALP00000018939 | GC | 4.16 × 10−9 | 0.11 | 3.68 | - | DOWN |
| Group Description | No. of Proteins | % Expression out of Caecum-Specific Proteins | Proteins |
|---|---|---|---|
| Drug metabolic processes and metabolism of exogenous substances | 12 | 22.48 | AKR1B1L, ALDH1A1, PRDX6, MPST, CBS, ADH1C, SELENBP1, SULT1E1, NDUFV2, FH, NDUFS1, ENO2 |
| Wound healing | 12 | 17.39 | GAL-1, FN1, AHNAK, CAPZA2, FHL2, CAPZA1, PLG, TNC, NMNAT3, AIMP1, COL5A1, POSTN |
| Extracellular matrix organisation and cell–matrix adhesion proteins | 14 | 13.52 | FN1, NID2, NID1, PRELP, TNC, PLG, COL5A1, POSTN, AIMP1, CAST, TNXB, PXN, CTTN, ACTN2 |
| Cellular response to stress | 8 | 11.59 | TXN, TUBB2B, PRDX6, CAPZA2, CAPZA1, ATOX1, ST13, TNXB |
| H2S metabolism | 4 | 8.71 | CBS, MPST, TST, SELENBP1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volf, J.; Rajova, J.; Babak, V.; Seidlerova, Z.; Rychlik, I. Detoxification, Hydrogen Sulphide Metabolism and Wound Healing Are the Main Functions That Differentiate Caecum Protein Expression from Ileum of Week-Old Chicken. Animals 2021, 11, 3155. https://doi.org/10.3390/ani11113155
Volf J, Rajova J, Babak V, Seidlerova Z, Rychlik I. Detoxification, Hydrogen Sulphide Metabolism and Wound Healing Are the Main Functions That Differentiate Caecum Protein Expression from Ileum of Week-Old Chicken. Animals. 2021; 11(11):3155. https://doi.org/10.3390/ani11113155
Chicago/Turabian StyleVolf, Jiri, Jana Rajova, Vladimir Babak, Zuzana Seidlerova, and Ivan Rychlik. 2021. "Detoxification, Hydrogen Sulphide Metabolism and Wound Healing Are the Main Functions That Differentiate Caecum Protein Expression from Ileum of Week-Old Chicken" Animals 11, no. 11: 3155. https://doi.org/10.3390/ani11113155
APA StyleVolf, J., Rajova, J., Babak, V., Seidlerova, Z., & Rychlik, I. (2021). Detoxification, Hydrogen Sulphide Metabolism and Wound Healing Are the Main Functions That Differentiate Caecum Protein Expression from Ileum of Week-Old Chicken. Animals, 11(11), 3155. https://doi.org/10.3390/ani11113155






