Emodin Improves Intestinal Health and Immunity through Modulation of Gut Microbiota in Mice Infected by Pathogenic Escherichia coli O1
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Culture and Diarrhea Modelling
2.2. Animals and Trial Groups
2.3. Sample Collection
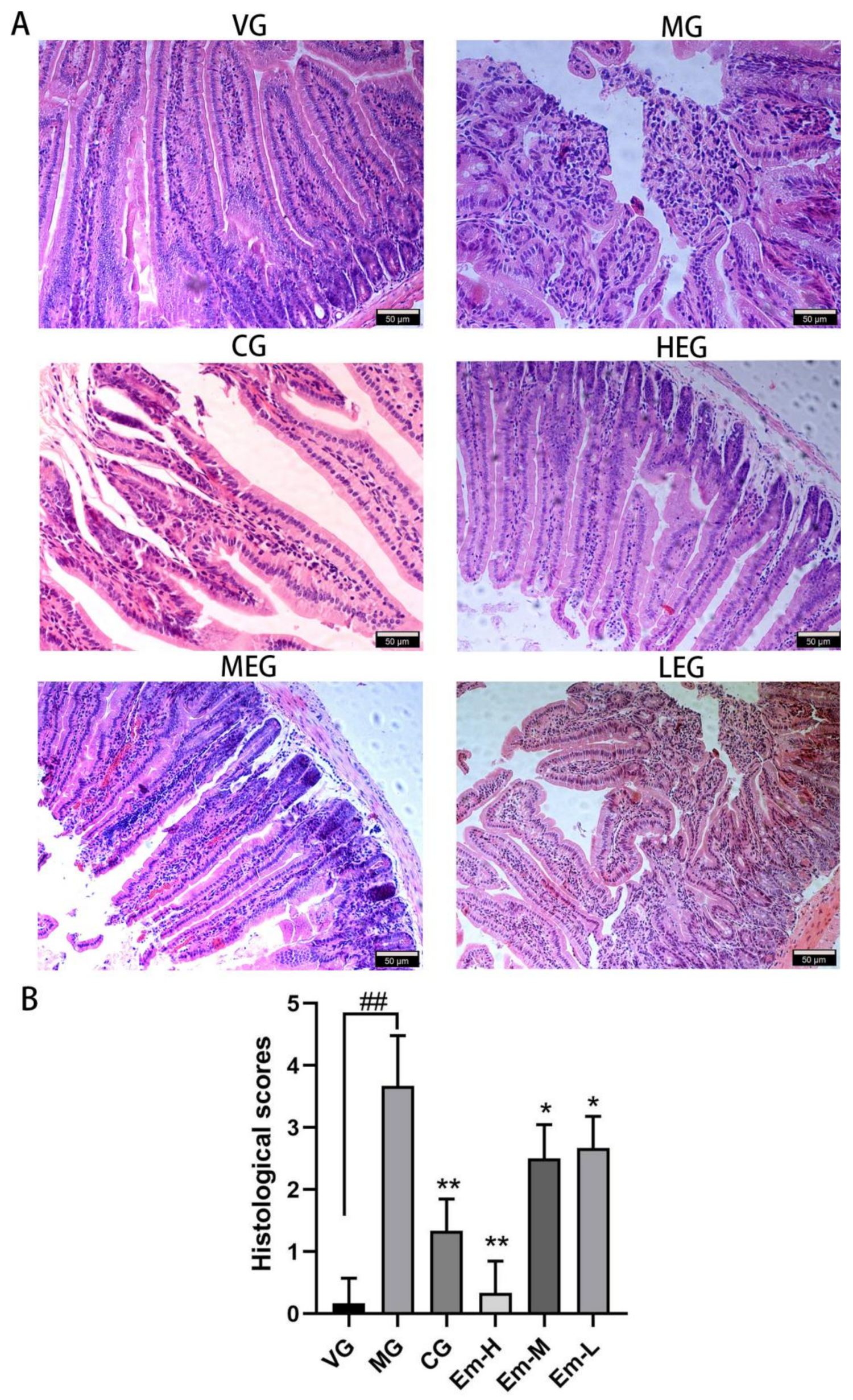
2.4. Histopathological Examination
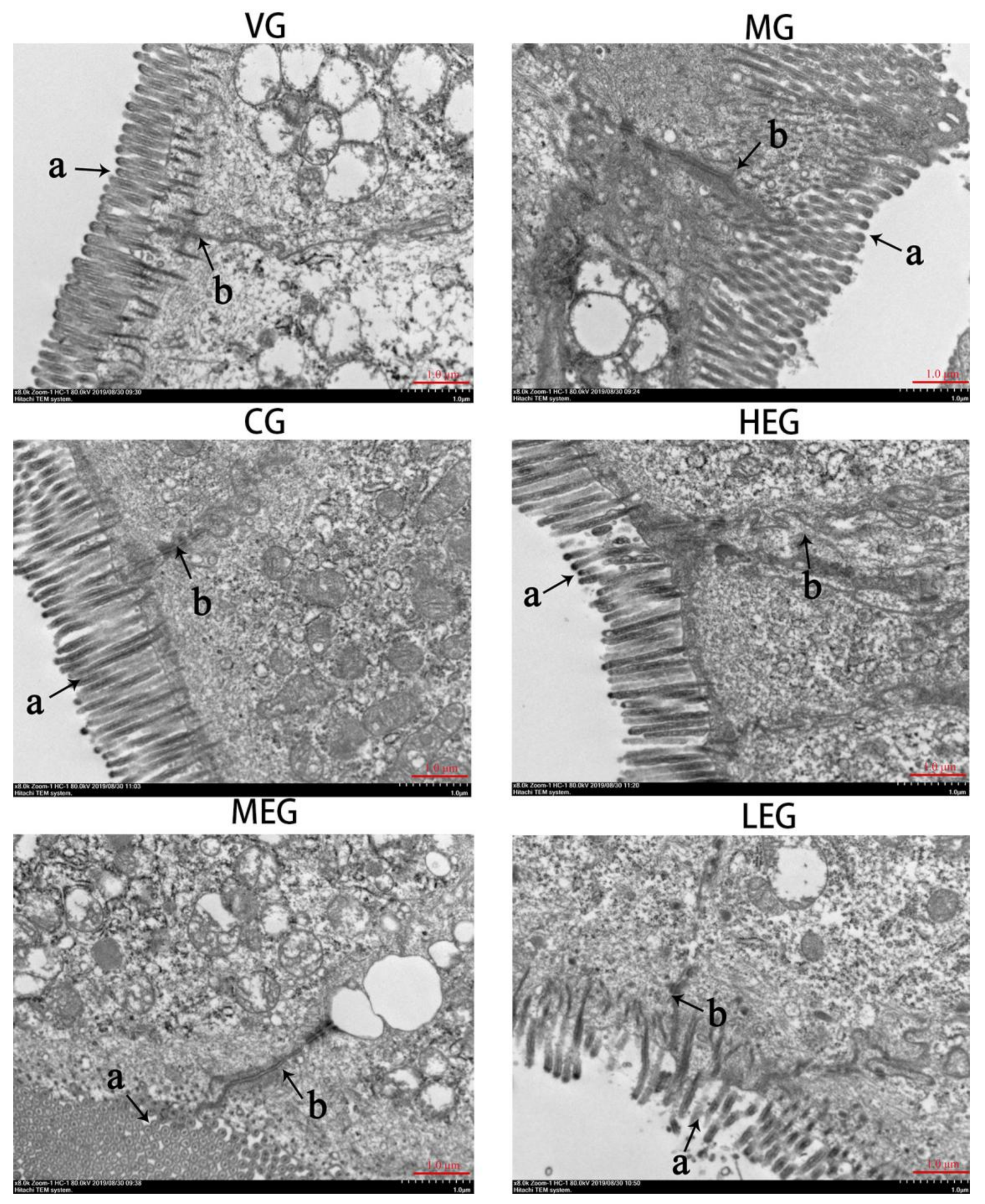
2.5. Transmission Electron Microscopy (TEM)
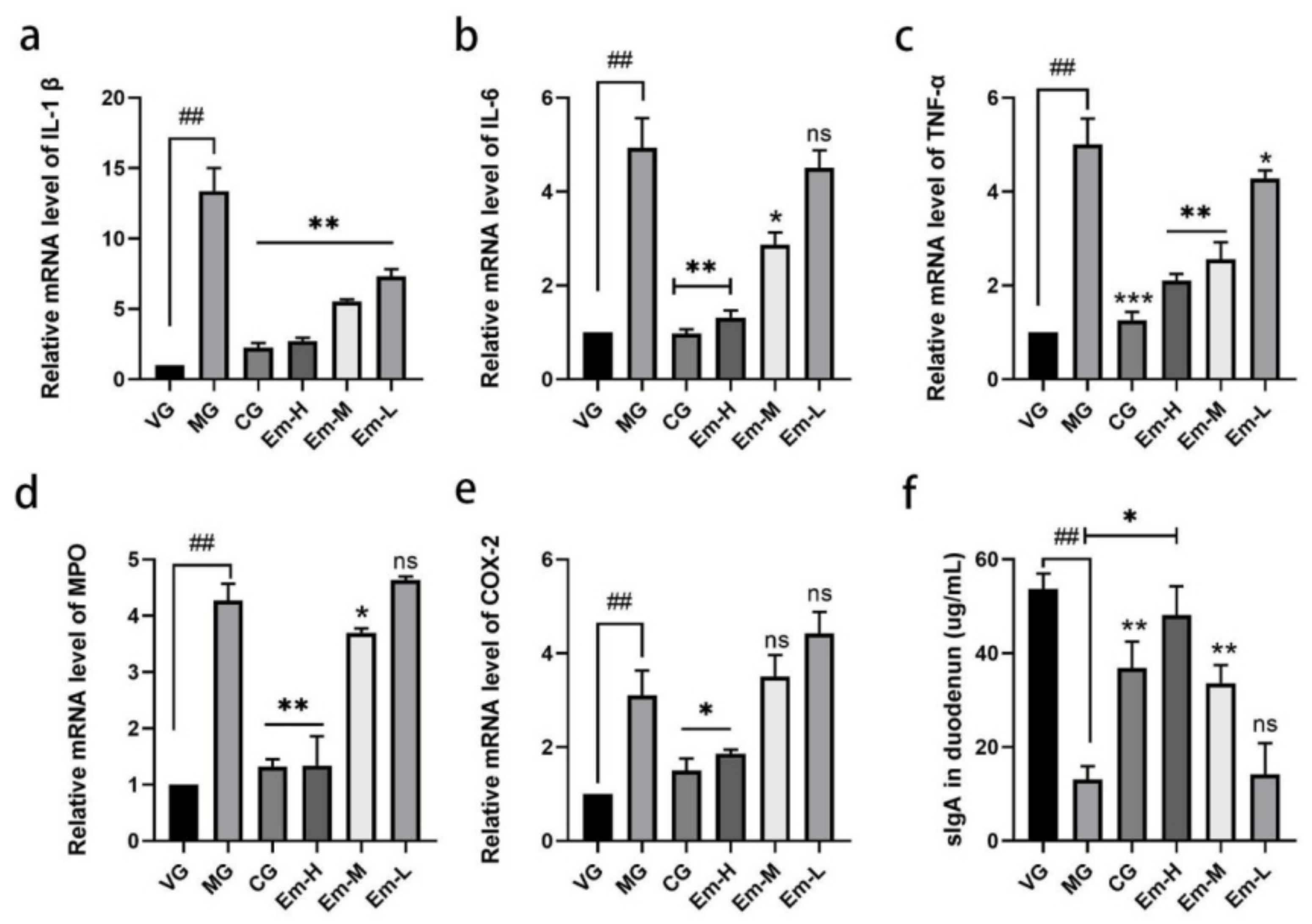
2.6. Real-Time PCR Assay and ELISA Assays
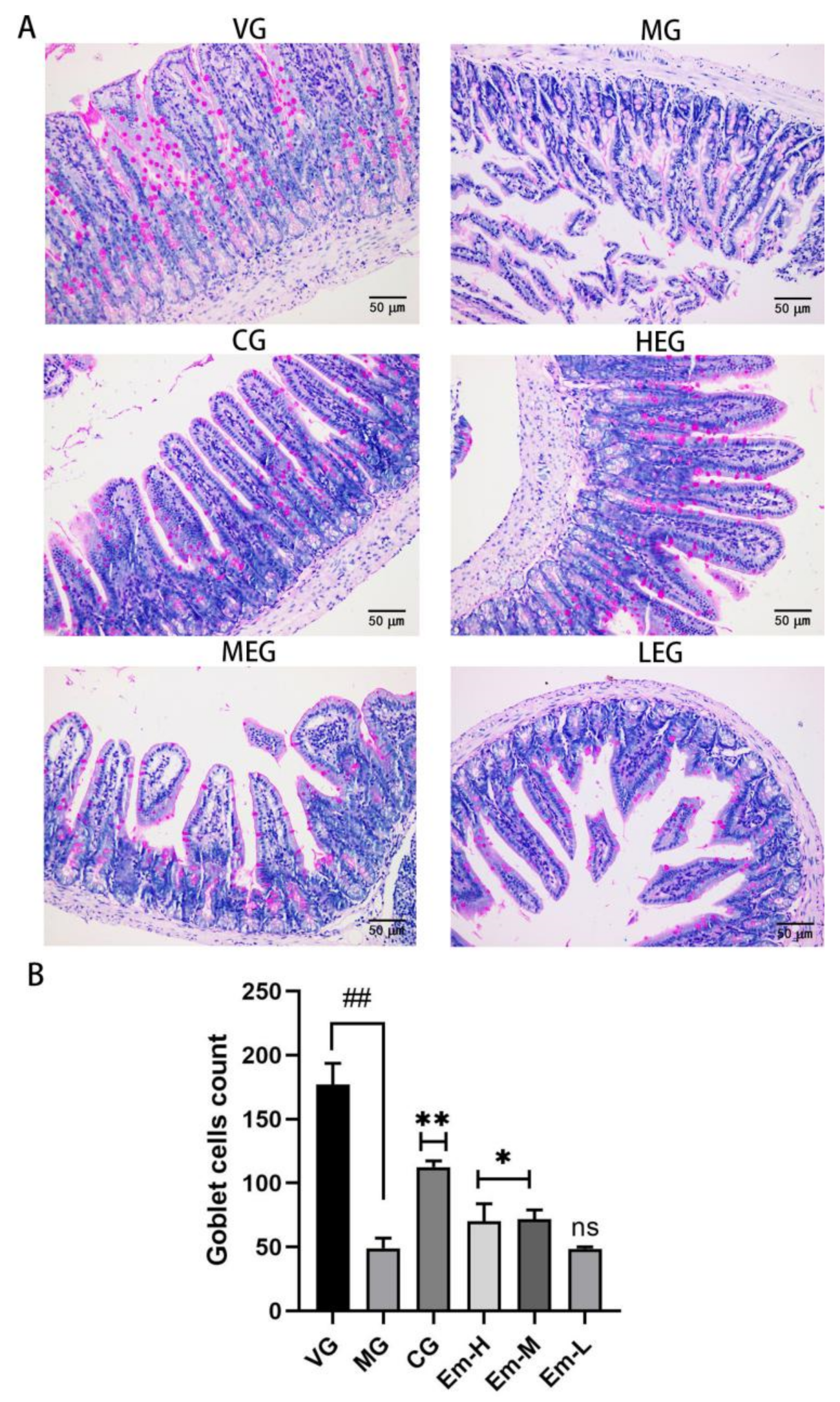
2.7. PAS Staining
2.8. Immunofluorescence
2.9. DNA Extraction and Hiseq Sequencing
2.10. 16S rRNA Bioinformatics Analysis and Statistics
3. Results
3.1. Histopathological Changes of Duodenum
3.2. Ultrastructural Changes in Duodenum
3.3. Cytokine Changes in Duodenum
3.4. Changes in Duodenal Goblet Cells
3.5. Changes of MUC-2 in the Duodenum
3.6. Changes in Intestinal Microbiota
3.6.1. Alpha Diversity Analysis
3.6.2. Beta Diversity Analysis
3.6.3. Taxonomic Composition of Intestinal Microbiota
3.6.4. LEFse Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Tojo, R.; Suárez, A.; Clemente, M.G.; de los Reyes-Gavilán, C.G.; Margolles, A.; Gueimonde, M.; Ruas-Madiedo, P. Intestinal microbiota in health and disease: Role of bifidobacteria in gut homeostasis. World J. Gastroenterol. 2014, 20, 15163–15176. [Google Scholar] [CrossRef]
- Tuddenham, S.; Sears, C.L. The intestinal microbiome and health. Curr. Opin. Infect. Dis. 2015, 28, 464–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, Y.F.; Hu, J.; Yan, X.H. Cross-talk between bile acids and intestinal microbiota in host metabolism and health. J. Zhejiang Univ. Sci. B 2015, 16, 436–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, K.; Decoffe, D.; Molcan, E.; Gibson, D.L. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 2012, 4, 1095–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sjögren, Y.M.; Tomicic, S.; Lundberg, A.; Böttcher, M.F.; Björkstén, B.; Sverremark-Ekström, E.; Jenmalm, M.C. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin. Exp. Allergy 2009, 39, 1842–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, C.; Neurath, M.F.; Wirtz, S. The intestinal microbiota in inflammatory bowel disease. ILAR J. 2015, 56, 192–204. [Google Scholar] [CrossRef] [Green Version]
- Sabatino, A.; Regolisti, G.; Cosola, C.; Gesualdo, L.; Fiaccadori, E. Intestinal microbiota in type 2 diabetes and chronic kidney disease. Curr. Diabetes Rep. 2017, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Keku, T.O.; Dulal, S.; Deveaux, A.; Jovov, B.; Han, X. The gastrointestinal microbiota and colorectal cancer. Am. J. Physiol.-Gastrointest. Liver Physiol. 2015, 308, G351–G363. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Adachi, K.; Sugiyama, T.; Shimozato, A.; Ebi, M.; Ogasawara, N.; Funaki, Y.; Goto, C.; Sasaki, M.; Kasugai, K. Association of intestinal microbiota with metabolic markers and dietary habits in patients with type 2 diabetes. Digestion 2016, 94, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Bin, P.; Tang, Z.; Liu, S.; Chen, S.; Xia, Y.; Liu, J.; Wu, H.; Zhu, G. Intestinal microbiota mediates Enterotoxigenic Escherichia coli-induced diarrhea in piglets. BMC Vet. Res. 2018, 14, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Gao, Y.; He, G.; Wang, Y.; Feng, B.; Hu, Y.; Mu, X.; Zhang, Y.; Dong, H. Escherichia coli O101-induced diarrhea develops gut microbial dysbiosis in rats. Exp. Ther. Med. 2019, 17, 824–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devriese, S.; Eeckhaut, V.; Geirnaert, A.; den Bossche, L.V.; Hindryckx, P.; de Wiele, T.V.; Van Immerseel, F.; Ducatelle, R.; De Vos, M.; Laukens, D. Reduced mucosa-associated Butyricicoccus activity in patients with ulcerative colitis correlates with aberrant claudin-1 expression. J. Crohn’s Colitis 2017, 11, 229–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Wang, R.; Liang, H.; Zhu, Q.; Liu, Y.; Yang, M.; Fang, Z.; Lin, Y.; Xu, S.; Feng, B.; et al. Enterococcus faecium NCIMB 10415 administration improves the intestinal health and immunity in neonatal piglets infected by enterotoxigenic Escherichia coli K88. J. Anim. Sci. Biotechnol. 2020, 72, 132–146. [Google Scholar] [CrossRef]
- Fleckenstein, J.M.; Hardwige, P.H.; Munson, G.P.; Rasko, D.A.; Sommerfelt, H.; Steinsland, H. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 2010, 12, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Tianwei, W.; Teng, K.; Liu, G.; Liu, Y.; Zhang, J.; Zhang, X.; Zhang, M.; Tao, Y.; Zhong, J. Lactobacillus reuteri HCM2 protects mice against Enterotoxigenic Escherichia coli through modulation of gut microbiota. Sci. Rep. 2018, 8, 17485. [Google Scholar]
- Gresse, R.; Chaucheyras-Durand, F.; Alain Fleury, M.; Van de Wiele, T.; Forano, E.; Blanquet-Diot, S. Gut microbiota dysbiosis in postweaning piglets: Understanding the keys to health. Trends Microbiol. 2017, 10, 851–873. [Google Scholar] [CrossRef] [PubMed]
- Nagy, B.; Fekete, P.Z. Enterotoxigenic Escherichia coli in veterinary medicine. Int. J. Med. Microbiol. 2005, 295, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Yin, J.; Duan, J.; Liu, G.; Zhu, X.; Chen, S.; Li, T.; Wang, S.; Tang, Y.; Hardwidge, P.R. Mouse intestinal innate immune responses altered by enterotoxigenic Escherichia coli (ETEC) infection. Microbes Infect. 2014, 16, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Chandler, D.; Mynott, T. Bromelain protects piglets from diarrhoea caused by oral challenge with K88 positive enterotoxigenic Escherichia coli. Gut 1998, 43, 196–202. [Google Scholar] [CrossRef] [Green Version]
- Ren, W.; Yin, J.; Xiao, H.; Chen, S.; Liu, G.; Tan, B.; Li, N.; Li, T.; Zent, B. Intestinal microbiota-derived GABA mediates interleukin-17 expression during enterotoxigenic Escherichia coli infection. Front. Immunol. 2016, 7, 685. [Google Scholar] [CrossRef] [Green Version]
- Ly, W.; Liu, C.; Ye, C.; Sun, J.; Tan, X.; Zhang, C.; Qu, Q.; Shi, D. Structural modulation of gut microbiota during alleviation of antibiotic-associated diarrhea with herbal formula. Int. J. Biol. Macromol. 2017, 105, 1622–1629. [Google Scholar]
- Solomon, S.L.; Oliver, K.B. Antibiotic resistance threats in the United States: Stepping back from the brink. Am. Fam. Physician 2014, 89, 938–941. [Google Scholar] [PubMed]
- Shan, T.; Wang, H.; Sha, L.; Bai, T.N.; Gong, J.H.; Jin, W.J.; Dai, L.L.; Ba, G.N.; Cho, S.B.; Fu, M.H. Protective effect and mechanisms of action of Mongolian medicine Sulongga-4 on pyloric ligation-induced gastroduodenal ulcer in rats. World J. Gastroentol. 2021, 27, 1770–1784. [Google Scholar]
- Sun, H.; Ye, Z.; Li, N.; Jin, F.; Yan, J.; Wu, K. Effect of emodin on T cell subsets in NOD mice with NaI-induced experimental autoimmune thyroiditis. Mol. Med. Rep. 2018, 18, 4303–4312. [Google Scholar] [CrossRef] [Green Version]
- Qu, K.; Shen, N.; Xu, X.; Su, H.; Wei, J.; Tai, M.; Meng, F.; Zhou, L.; Zhang, Y.; Liu, C. Emodin induces human T cell apoptosis in vitro by ROS-mediated endoplasmic reticulum stress and mitochondrial dysfunction. Acta Pharmacol. Sin. 2013, 34, 1217–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Z.; Wu, K.; Jin, F.; Xu, T.; Li, N.; Huang, J. Effect of emodin on intestinal flora in the treatment of iodine-induced thyroiditis in NOD mice. Int. J. Clin. Exp. Med. 2020, 13, 1493–1500. [Google Scholar]
- Daly, S.M.; Elmore, B.O.; Kavanaugh, J.S.; Triplett, K.D.; Figueroa, M.; Raja, H.A.; El-Elimat, T.; Crosby, H.A.; Femling, J.K.; Cech, N.B.; et al. ω-Hydroxyemodin limits staphylococcus aureus quorum sensing-mediated pathogenesis and inflammation. Antimicrob. Agents Chemother. 2015, 59, 2223–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrera-Benitez, N.E.; Pérez-Roth, E.; Casula, M.; Ramos-Nuez, A.; Ríos-Luci, C.; Rodríguez-Gallego, C.; Sologuren, I.; Jakubkiene, V.; Slutsky, A.S.; Padrón, J.M.; et al. Anti-inflammatory activity of a novel family of aryl ureas compounds in an endotoxin-induced airway epithelial cell injury model. PLoS ONE 2012, 7, e48468. [Google Scholar] [CrossRef]
- Kim, Y.S.; Lee, Y.M.; Oh, T.I.; Shin, D.H.; Kim, G.H.; Kan, S.Y.; Kang, H.; Kim, J.H.; Kim, B.M.; Yim, W.J.; et al. Emodin sensitizes hepatocellular carcinoma cells to the anti-cancer effect of sorafenib through suppression of cholesterol metabolism. Int. J. Mol. Sci. 2018, 19, 3127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, J.-W.; Zhang, Y.; Yu, M.-S.; Gu, M.; Xu, J.; Usman, A.; Ji, F. Emodin alleviates intestinal mucosal injury in rats with severe acute pancreatitis via the caspase-1 inhibition. Hepatobiliary Pancreat. Dis. Int. 2017, 16, 431–436. [Google Scholar] [CrossRef]
- Jia, Z.; Chen, A.; Bao, F.; He, M.; Gao, S.; Xu, J.; Zhang, X.; Niu, P.; Wang, C. Effect of nisin on microbiome-brain-gut axis neurochemicals by Escherichia coli-induced diarrhea in mice. Microb. Pathog. 2018, 119, 65–71. [Google Scholar] [CrossRef]
- Li, Y.; Guo, R.; Zhang, M.; Chen, P.; Li, J.; Sun, Y. Protective effect of emodin on intestinal epithelial tight junction barrier integrity in rats with sepsis induced by cecal ligation and puncture. Exp. Ther. Med. 2020, 19, 3521–3530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.N.; Zeng, Z.G.; Wang, H.N.; Yang, T.; Zhang, P.J.; Li, Y.L.; Zhang, A.Y.; Fan, W.Q.; Zhang, Y.; Yang, X.; et al. An effective method for isolation of DNA from pig feces and comparison of five different methods. J. Microbiol. Methods 2008, 75, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Huasai, S.; Wang, C.; Zhang, J.; Lv, W.; Xirnud, J.; Chen, A. Comparative analysis of fecal microbiota of grazing Mongolian cattle from different regions in Inner Mongolia, China. Animals 2021, 7, 1938. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, J.; Han, H.; Li, Y.; Liu, Z.J.; Zhao, Y.R.; Fang, R.J.; Huang, X.G.; Zheng, J.; Ren, W.K.; Wu, F.; et al. Lysine restriction affects feed intake and amino acid metabolism via gut microbiome in piglets. Cell. Physiol. Biochem. 2017, 44, 1749–1761. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartfield, D.; Turner, J.; Huynh, H.; Lidman, P.; Chaba, T.; Lacson, A. The role of histopathology in diagnosing protracted diarrhea of infancy. Fetal Pediatr. Pathol. 2010, 29, 144–157. [Google Scholar] [CrossRef]
- Simmerson, S.M.; Armstrong, P.J.; Wünschmann, A.; Jessen, C.R.; Crews, L.J.; Washabau, R.J. Clinical features, intestinal histopathology, and outcome in protein-losing enteropathy in Yorkshire Terrier dogs. J. Vet. Int. Med. 2014, 28, 331–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Piao, X.S.; Thacker, P.A.; Zeng, Z.K.; Li, P.F.; Wang, D.; Kim, S.W. Chito-oligosaccharide reduces diarrhea incidence and attenuates the immune response of weaned pigs challenged with Escherichia coli K88. J. Anim. Sci. 2010, 88, 3871–3879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W. Metabolism of Emodin in Liver and Intestinal and Its Gender-Differences. Ph.D. Dissertation, Southern Medical University, Guangzhou, China, 2010. [Google Scholar]
- Basu, S.; Ghosh, A.; Hazra, B. Evaluation of the antibacterial activity of Ventilago madraspatana Gaertn., Rubia cordifolia Linn. and Lantana camara Linn.: Isolation of emodin and physcion as active antibacterial agents. Phytother. Res. 2005, 19, 888–894. [Google Scholar] [CrossRef]
- Turner, J.R. Molecular basis of epithelial barrier regulation: From basic mechanisms to clinical application. Am. J. Pathol. 2006, 169, 1901–1909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piel, C.; Montagne, L.; Sève, B.; Lallès, J.P. Increasing digesta viscosity using carboxymethylcellulose in weaned piglets stimulates ileal goblet cell numbers and maturation. J. Nutr. 2005, 135, 86–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, F.S.; Romero-Calvo, I.; Mascaraque, C.; Martínez-Augustin, O. Intestinal inflammation and mucosal barrier function. Inflamm. Bowel Dis. 2014, 20, 2394–2404. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Mu, J.K.; Li, W.Q.; Tong, Z.H.; Li, J.; Zheng, S.Y. Effects of early enteral nutrition on immune function of severe acute pancreatitis patients. World J. Gastroentol. 2013, 19, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; DeStefano, J.; Meier, M.F.; Muegge, B.D.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef]
- Toki, S.; Kagaya, S.; Shinohara, M.; Wakiguchi, H.; Matsumoto, T.; Takahata, Y.; Morimatsu, F.; Saito, H.; Matsumoto, K. Lactobacillus rhamnosus GG and Lactobacillus casei suppress Escherichia coli-induced chemokine expression in intestinal epithelial cells. Int. Arch. Allergy Immunol. 2009, 148, 45–58. [Google Scholar] [CrossRef]
- Li, Y.; Liao, Q.; Lin, M.; Zhong, D.; Wei, L.; Han, B.; Miao, H.; Yao, M.; Xie, Z. An integrated metabonomics and microbiology analysis of host-microbiota metabolic interactions in rats with Coptis chinensis-induced diarrhea. RSC Adv. 2015, 5, 79329–79341. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, Y.H.; Zhou, D.; Wu, Q.; Song, D.; Dicksyed, J.; Wang, J.F. Oral administration of a select mixture of Bacillus probiotics affects the gut microbiota and goblet cell function following Escherichia coli challenge in newly weaned pigs of genotype MUC4 that are supposed to be enterotoxigenic E. coli F4ab/ac receptor negative. Appl. Environ. Microbiol. 2017, 83, e02747-16. [Google Scholar]
- Andreasen, A.S.; Larsen, N.; Pedersen-Skovsgaard, T.; Berg, R.M.G.; Møller, K.; Svendsen, K.D.; Jakobsen, M.; Pedersen, B.K. Effects of Lactobacillus acidophilus NCFM on insulin sensitivity and the systemic inflammatory response in human subjects. Br. J. Nutr. 2010, 104, 1831–1838. [Google Scholar] [CrossRef] [Green Version]
- Hakansson, A.; Molin, G. Gut microbiota and inflammation. Nutrients 2011, 3, 637–682. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, L.; Zhai, Z.; Zhao, W.; Ding, J.; Dai, R.; Sun, T.; He, M. Porcine epidemic diarrhea virus infection induced the unbalance of gut microbiota in piglets. Curr. Microbiol. 2015, 71, 643–649. [Google Scholar] [CrossRef]
- Tropini, C.; Moss, E.L.; Merrill, B.D.; Ng, K.M.; Higginbottom, S.K.; Casavant, E.P.; Gonzalez, C.G.; Fremin, B.; Bouley, D.M.; Elias, J.E.; et al. Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell 2018, 173, 1742–1754. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wu, Y.; Xu, Z.; Chen, J.; Li, Y.; Hanjing, X.; Zhang, X.; Yuan, J. Effects of Hetiao Jianpi Decoction on intestinal injury and repair in rats with antibiotic-associated diarrhea. Med. Sci. Monit. 2020, 26, e921745-1–e921745-10. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef]
- Xiang, H.; Cao, F.; Ming, D.; Zheng, Y.; Dong, X.; Zhong, X.; Mu, D.; Li, B.; Zhong, L.; Cao, J.; et al. Aloe-emodin inhibits Staphylococcus aureus biofilms and extracellular protein production at the initial adhesion stage of biofilm development. Appl. Microbiol. Biotechnol. 2017, 101, 6671–6681. [Google Scholar] [CrossRef]
- Li, X.; Shan, C.; Wu, Z.; Yu, H.; Yang, A.; Tan, B. Emodin alleviated pulmonary inflamma tion in rats with LPS-induced acute lung injury through inhibiting the mTOR/HIF-1α/VEGF signaling pathway. Inflamm. Res. 2020, 69, 365–373. [Google Scholar] [CrossRef]
- Saunders, I.T.; Mir, H.; Kapur, N.; Singh, S. Emodin inhibits colon cancer by altering BCL-2 family proteins and cell survival pathways. Cancer Cell Int. 2019, 19, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, S.; Wang, K.; Yu, W.; Sun, B.; Schwarz, W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antivir. Res. 2011, 90, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Qiang, F.; Chao, D.; Di, W.; Guoqian, Z.; Bo, Y.; Lina, Y. Amelioration of hypoxia and LPS-induced intestinal epithelial barrier dysfunction by emodin through the suppression of the NF-kappaB and HIF-1alpha sig naling pathways. Int. J. Mol. Med. 2014, 34, 1629–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]

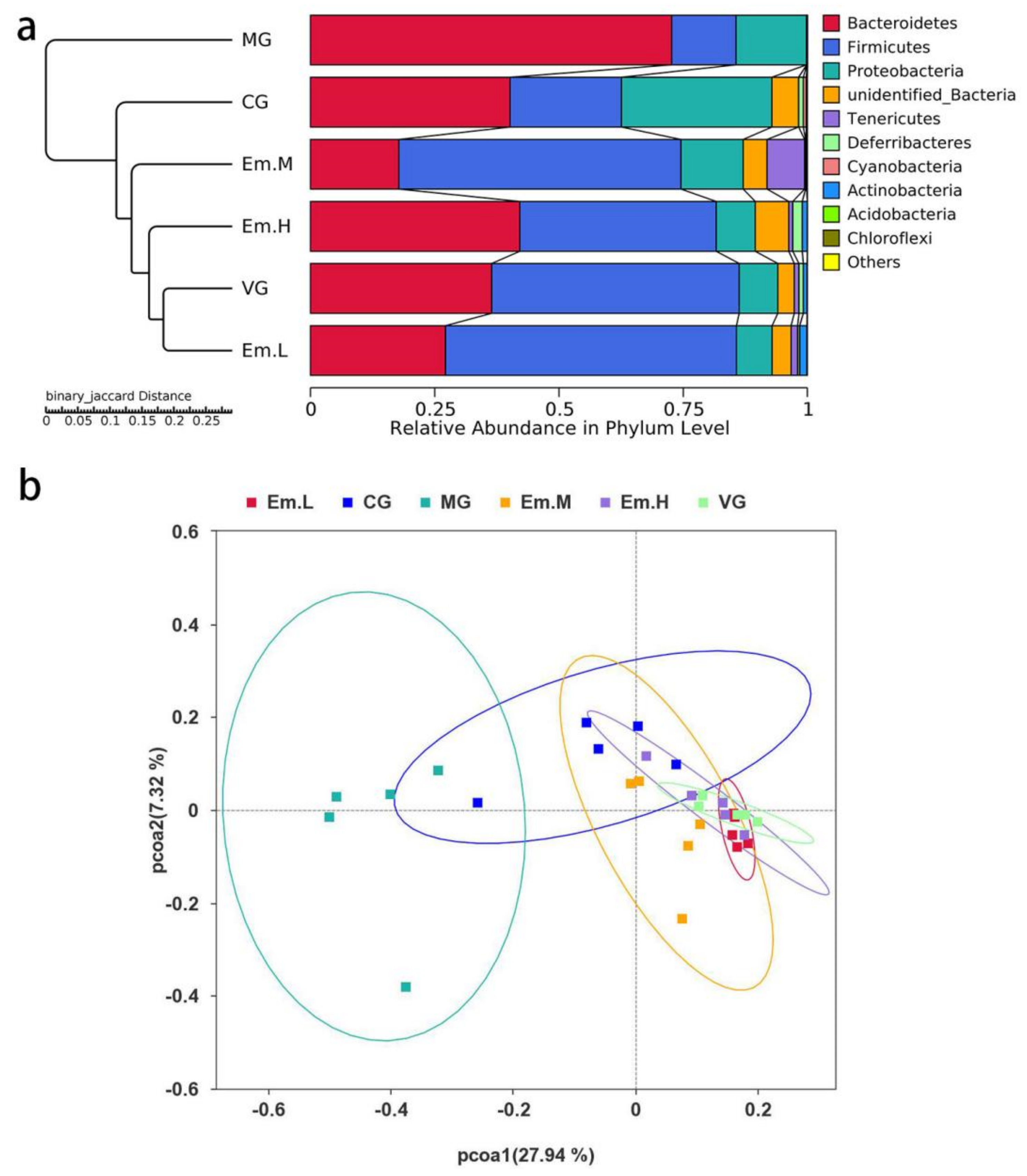
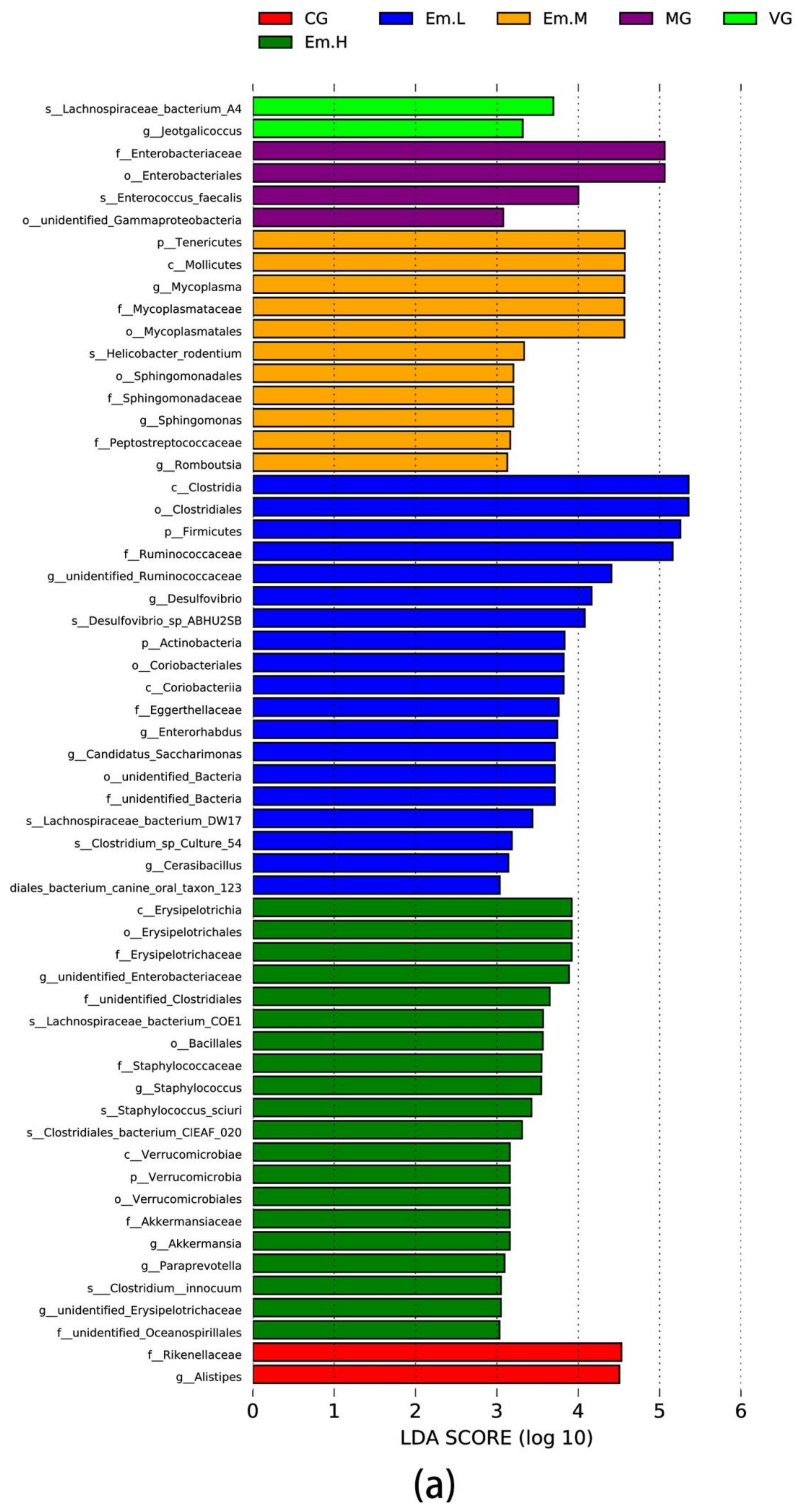
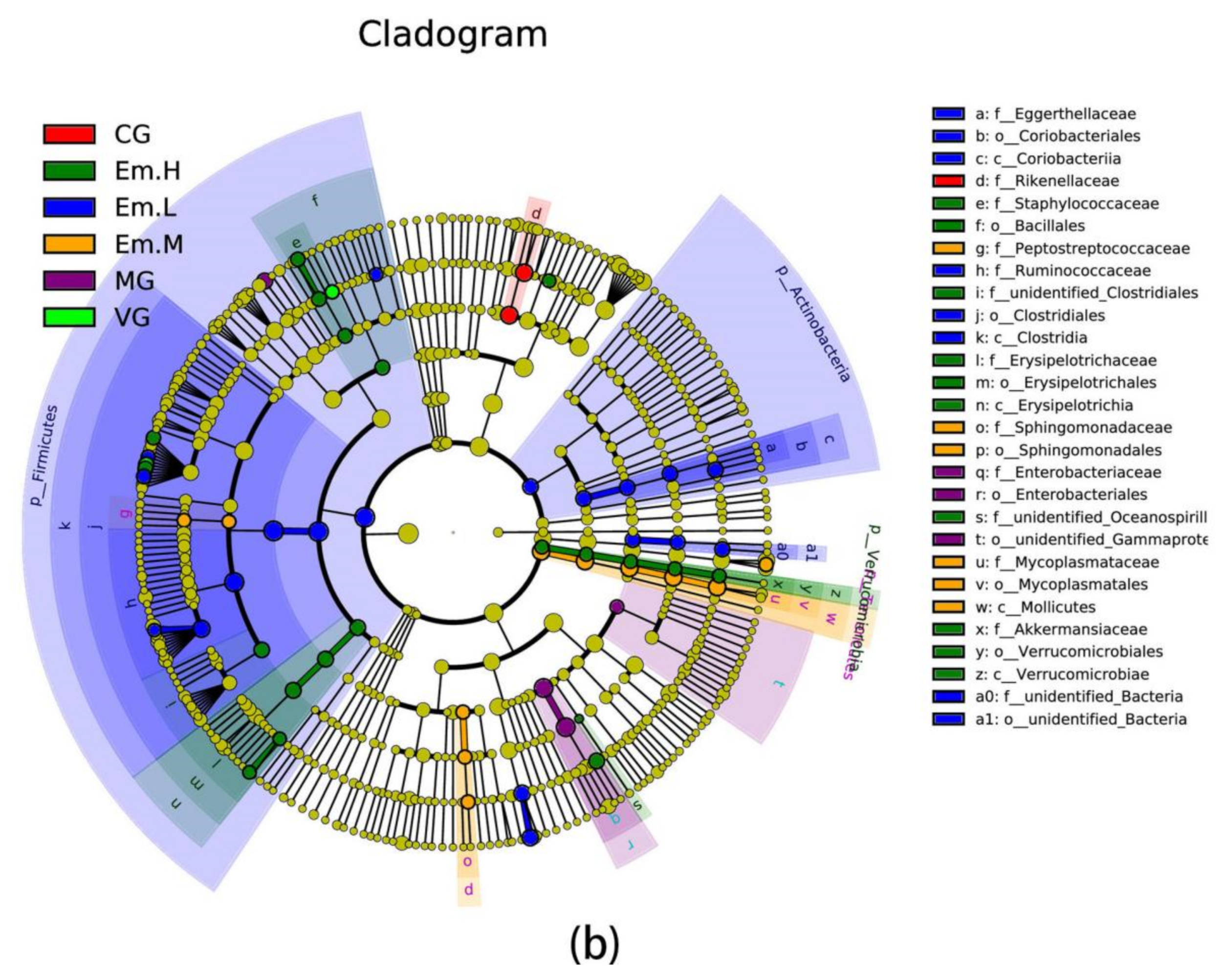
| Group | Community Diversity | Community Richness | ||||
|---|---|---|---|---|---|---|
| Shannon | Simpon | Chao1 | ACE | Observed_Species | Goods_Coverage | |
| VG | 6.36 ± 0.18 a | 0.44 ± 0.12 c | 524.52 ± 44.31 a | 515.31 ± 18.84 a | 463.60 ± 19.68 a | 0.998 ± 0.0002 |
| MG | 1.70 ± 0.18 c | 0.96 ± 0.13 a | 216.56 ± 16.88 c | 233.26 ± 14.09 c | 174.80 ± 16.52c | 0.998 ± 0.0002 |
| CG | 4.11 ± 0.45 b | 0.87 ± 0.08 b | 370.63 ± 41.56 b | 386.65 ± 15.44 b | 325.40 ± 18.38 b | 0.998 ± 0.0002 |
| Em-H | 6.03 ± 0.35 a | 0.40 ± 0.06 c | 527.11 ± 27.30 a | 521.03 ± 23.90 a | 460.00 ± 26.04 a | 0.998 ± 0.0002 |
| Em-M | 5.56 ± 0.31 a | 0.46 ± 0.04 c | 487.61 ± 36.78 a | 476.59 ± 37.01 a | 412.80 ± 37.08 a | 0.998 ± 0.0002 |
| Em-L | 6.45 ± 0.28 a | 0.46 ± 0.09 c | 566.27 ± 24.78 a | 550.91 ± 21.23 a | 503.40 ± 18.42 a | 0.998 ± 0.0002 |
| Relative Abundance (%) | Groups | |||||
|---|---|---|---|---|---|---|
| VG | MG | CG | Em-H | Em-H | Em-H | |
| Bacteroidetes | 0.93 ± 0.04 a | 0.09 ± 0.07 c | 0.32 ± 0.05 b | 0.92 ± 0.04 a | 0.42 ± 0.16 b | 0.27 ± 0.01 c |
| Firmicutes | 0.82 ± 0.09 a | 0.24 ± 0.07 d | 0.31 ± 0.07 d | 0.10 ± 0.06 b | 0.50 ± 0.05 c | 0.30 ± 0.12 d |
| Proteobacteria | 0.11 ± 0.02 c,d | 0.63 ± 0.05 a | 0.19 ± 0.06 c | 0.07 ± 0.01 d | 0.56 ± 0.01 d | 0.32 ± 0.06 b |
| Verrucomicrobia | 0.23 ± 0.07 a | 0.05 ± 0.00 c | 0.10 ± 0.00 bc | 0.15 ± 0.02 b | 0.05 ± 0.00 c | 0.03 ± 0.01 c |
| Relative Abundance (%) | Groups | |||||
|---|---|---|---|---|---|---|
| VG | MG | CG | Em-H | Em-M | Em-L | |
| Bacteroidetes | 0.85 ± 0.05 a | 0.04 ± 0.01 c | 0.22 ± 0.06 d | 0.45 ± 0.03 b | 0.35 ± 0.00 c | 0.25 ± 0.04 d |
| Clostridiales | 0.03 ± 0.01 e | 0.67 ± 0.11 a | 0.34 ± 0.01 c | 0.26 ± 0.02 d | 0.48 ± 0.03 b | 0.53 ± 0.01 b |
| Enterobacteriales | 0.01 ± 0.00 c | 0.35 ± 0.03 a | 0.01 ± 0.00 c | 0.01 ± 0.00 c | 0.01 ± 0.00 c | 0.11 ± 0.00 b |
| Lactobacillus | 0.33 ± 0.03 a | 0.03 ± 0.01 c | 0.15 ± 0.00 b | 0.05 ± 0.00 c | 0.03 ± 0.00 c | 0.02 ± 0.00 c |
| Verrucomicrobiales | 0.23 ± 0.07 a | 0.05 ± 0.00 c | 0.11 ± 0.00 bc | 0.15 ± 0.02 b | 0.04 ± 0.00 c | 0.03 ± 0.01 c |
| Desulfovibrionales | 0.07 ± 0.00 a | 0.18 ± 0.00 a | 0.19 ± 0.00 a | 0.03 ± 0.00 a | 0.04 ± 0.00 a | 0.05 ± 0.00 a |
| Campylobacterales | 0.03 ± 0.00 a | 0.05 ± 0.00 a | 0.00 ± 0.00 a | 0.06 ± 0.00 a | 0.04 ± 0.00 a | 0.03 ± 0.00 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.; Wang, C.; Han, A.; Tian, Y.; Ren, S.; Lv, W.; Chen, A.; Zhang, J. Emodin Improves Intestinal Health and Immunity through Modulation of Gut Microbiota in Mice Infected by Pathogenic Escherichia coli O1. Animals 2021, 11, 3314. https://doi.org/10.3390/ani11113314
Gao R, Wang C, Han A, Tian Y, Ren S, Lv W, Chen A, Zhang J. Emodin Improves Intestinal Health and Immunity through Modulation of Gut Microbiota in Mice Infected by Pathogenic Escherichia coli O1. Animals. 2021; 11(11):3314. https://doi.org/10.3390/ani11113314
Chicago/Turabian StyleGao, Ruijuan, Chunjie Wang, Aricha Han, Yanping Tian, Shunan Ren, Wenting Lv, Aorigele Chen, and Jian Zhang. 2021. "Emodin Improves Intestinal Health and Immunity through Modulation of Gut Microbiota in Mice Infected by Pathogenic Escherichia coli O1" Animals 11, no. 11: 3314. https://doi.org/10.3390/ani11113314
APA StyleGao, R., Wang, C., Han, A., Tian, Y., Ren, S., Lv, W., Chen, A., & Zhang, J. (2021). Emodin Improves Intestinal Health and Immunity through Modulation of Gut Microbiota in Mice Infected by Pathogenic Escherichia coli O1. Animals, 11(11), 3314. https://doi.org/10.3390/ani11113314






