A Matrilineal Study on the Origin and Genetic Relations of the Ecuadorian Pillareño Creole Pig Population through D-Loop Mitochondrial DNA Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Ethics Statement
2.3. DNA Extraction and Amplification
2.4. Molecular D-Loop Analysis
2.4.1. Mitochondrial Genetic Diversity and Differentiation
2.4.2. Genealogical Relationships between Haplotypes
3. Results
3.1. Sequence Analysis, Genetic Diversity, and Differentiation
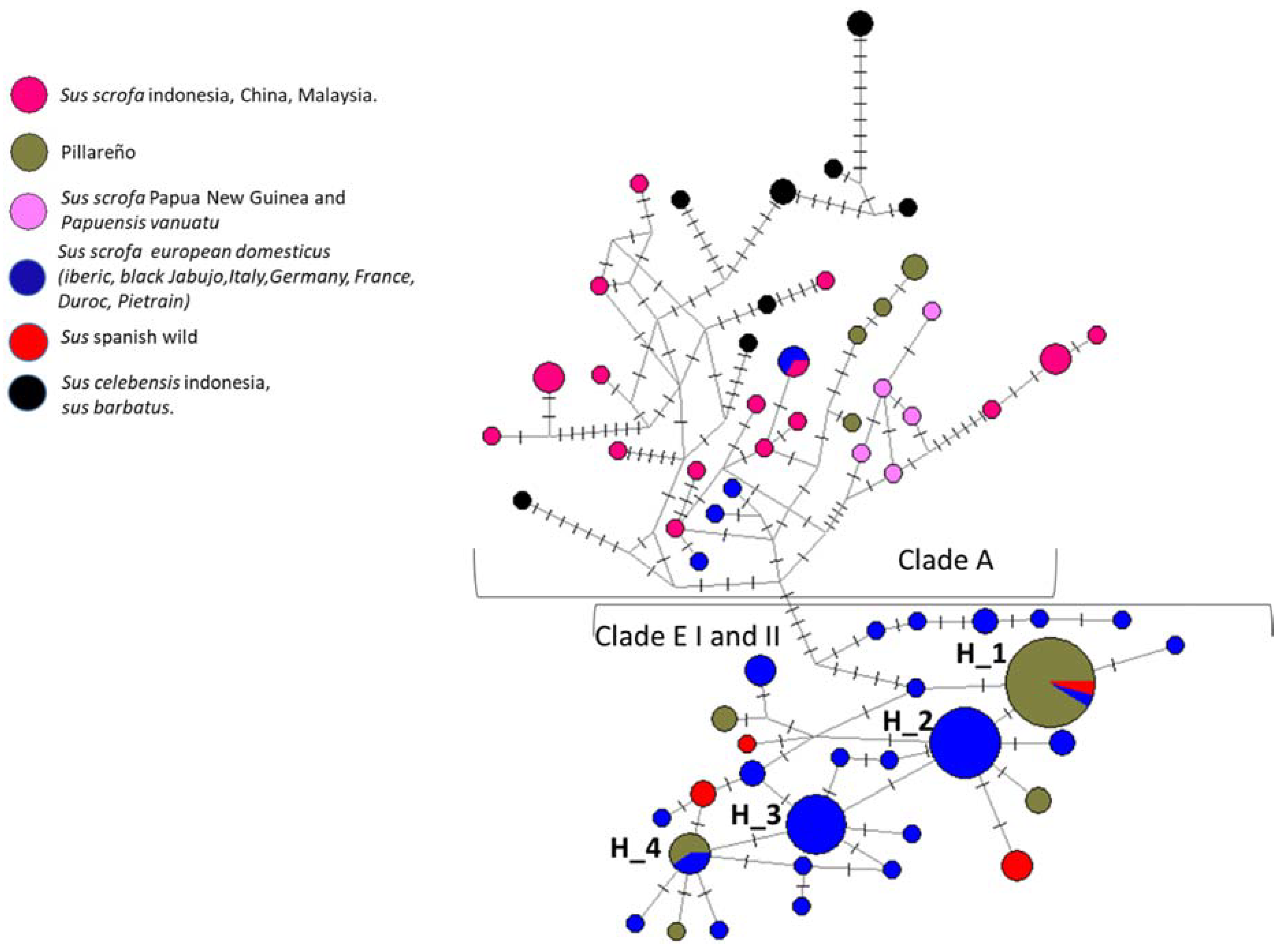
3.2. Haplotype Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casquet, J.; Thebaud, C.; Gillespie, R.G. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Mol. Ecol. Resour. 2012, 12, 136–141. [Google Scholar] [CrossRef]
- Ursing, B.M.; Arnason, U. The complete mitochondrial DNA sequence of the pig (Sus scrofa). J. Mol. Evol. 1998, 47, 302–306. [Google Scholar] [CrossRef]
- Watanobe, T.; Ishiguro, N.; Okumura, N.; Nakano, M.; Matsui, A.; Hongo, H.; Ushiro, H. Ancient mitochondrial DNA reveals the origin of Sus scrofa from Rebun Island, Japan. J. Mol. Evol. 2001, 52, 281–289. [Google Scholar] [CrossRef]
- Canales Vergara, A.M.; Landi, V.; Delgado Bermejo, J.V.; Martínez, A.; Cervantes Acosta, P.; Pons Barro, Á.; Bigi, D.; Sponenberg, P.; Helal, M.; Hossein Banabazi, M. Tracing worldwide turkey genetic diversity using D-loop sequence mitochondrial DNA analysis. Animals 2019, 9, 897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M. Phylogeny. fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Schneider, S.; Roessli, D.; Excoffier, L. Arlequin: A Software for Population Genetics Data Analysis; User Manual Ver. 2.000; Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva: Geneva, Switzerland, 2000; Volume 2, pp. 2496–2497. [Google Scholar]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Karhunen, M.; Ovaskainen, O. Estimating population-level coancestry coefficients by an admixture F model. Genetics 2012, 192, 609–617. [Google Scholar] [CrossRef] [Green Version]
- Giuffra, E.; Kijas, J.; Amarger, V.; Carlborg, Ö.; Jeon, J.-T.; Andersson, L. The origin of the domestic pig: Independent domestication and subsequent introgression. Genetics 2000, 154, 1785–1791. [Google Scholar] [CrossRef]
- Kijas, J.; Andersson, L. A phylogenetic study of the origin of the domestic pig estimated from the near-complete mtDNA genome. J. Mol. Evol. 2001, 52, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Toalombo, P.; Camacho, C.; Buenaño, R.; Jiménez, S.; Navas-González, F.; Landi, V.; Delgado, J. Efecto socioeconómico sobre las características fanerópticas de gallinas autóctonas de Ecuador. Arch. Zootec. 2019, 68, 416–421. [Google Scholar] [CrossRef] [Green Version]
- Lucas, L.S.; Lanari, M.; Oyarzabal, M. Phenotypic characterization of the goat population of Santa Elena province (Ecuador). Arch. Zootec. 2020, 69, 22–29. [Google Scholar]
- Estupiñan, K.; Barba, C.; Martínez, A.; Delgado, J. Caracterización genética del porcino Criollo de Ecuador. Arch. Zootec. 2020, 69, 384–388. [Google Scholar] [CrossRef]
- Bowen, B.; Grant, W.S. Phylogeography of the sardines (Sardinops spp.): Assessing biogeographic models and population histories in temperate upwelling zones. Evolution 1997, 51, 1601–1610. [Google Scholar] [CrossRef]
- Mock, K.E.; Theimer, T.; Rhodes, O., Jr.; Greenberg, D.; Keim, P. Genetic variation across the historical range of the wild turkey (Meleagris gallopavo). Mol. Ecol. 2002, 11, 643–657. [Google Scholar] [CrossRef]
- Guan, X.; Silva, P.; Gyenai, K.; Xu, J.; Geng, T.; Smith, E. Mitochondrial DNA-based analyses of relatedness among Turkeys, Meleagris gallopavo. Biochem. Genet. 2015, 53, 29–41. [Google Scholar] [CrossRef]
- Alves, E.; Ovilo, C.; Rodriguez, M.; Silio, L. Mitochondrial DNA sequence variation and phylogenetic relationships among Iberian pigs and other domestic and wild pig populations. Anim. Genet. 2003, 34, 319–324. [Google Scholar] [CrossRef]
- Lemus-Flores, C. Diversidad genética del cerdo criollo mexicano. Rev. Comput. Prod. Porc. 2008, 15, 33–40. [Google Scholar]
- Ramos-Onsins, S.E.; Rozas, J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 2002, 19, 2092–2100. [Google Scholar] [CrossRef] [Green Version]
- Rodero Serrano, E.; Rodero Franganillo, A.; Delgado-Bermejo, J. Primitive andalusian livestock and their implications in the discovery of America. Arch. Zootec. 1992, 41, 383–400. [Google Scholar]
- Jones, G. Genetic aspects of domestication, common breeds and their origin. In The Genetics of the Pig; CABI: Wallingford, UK, 1998; pp. 17–50. [Google Scholar]
- Vargas, J.; Velázquez, F.; Chacón, E. Estructura y relaciones genéticas del cerdo criollo de Ecuador. Rev. Electron. Vet. 2015, 16, 1–11. [Google Scholar]
- Fang, M.; Andersson, L. Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication. Proc. R. Soc. B Biol. Sci. 2006, 273, 1803–1810. [Google Scholar] [CrossRef] [Green Version]
- Valente, M.J.; Carvalho, A.F. Zooarchaeology in the Neolithic and Chalcolithic of southern Portugal. Environ. Archaeol. 2014, 19, 226–240. [Google Scholar] [CrossRef]
- Padilla-Jacobo, G.; Cano-Camacho, H.; López-Zavala, R.; Cornejo-Pérez, M.E.; Zavala-Páramo, M.G. Evolutionary history of Mexican domesticated and wild Meleagris gallopavo. Genet. Sel. Evol. 2018, 50, 19. [Google Scholar] [CrossRef] [Green Version]
- Thornton, E.K.; Emery, K.F.; Steadman, D.W.; Speller, C.; Matheny, R.; Yang, D. Earliest Mexican turkeys (Meleagris gallopavo) in the Maya region: Implications for pre-Hispanic animal trade and the timing of turkey domestication. PLoS ONE 2012, 7, e42630. [Google Scholar]
- Justo, L. El cerdo. Historia de un elemento esencial de la cultura castellana en la conquista y colonización de América (siglo XVI). Anu. Estud. Am. 1996, 53, 13–35. [Google Scholar]
| Nucleotide Positions | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotypes | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 4 | 4 | 4 | 5 | 5 | ||||||
| 3 | 6 | 6 | 8 | 8 | 0 | 0 | 1 | 3 | 3 | 7 | 7 | 9 | 3 | 3 | 5 | 5 | 6 | 8 | 4 | 0 | 0 | 5 | 1 | 3 | ||
| 3 | 5 | 6 | 0 | 7 | 1 | 9 | 4 | 7 | 8 | 0 | 1 | 7 | 6 | 9 | 1 | 8 | 3 | 0 | 7 | 0 | 9 | 8 | 7 | 2 | ||
| AJJ002189 | C | T | G | A | G | C | C | A | T | T | T | T | T | T | C | A | C | C | T | C | A | C | A | T | A | |
| H_1 | CPECU: 31 | . | C | . | . | A | T | T | G | C | C | C | . | C | C | . | G | . | T | . | T | . | T | G | C | G |
| H_2 | CPECU: 13, 21 | T | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . |
| H_3 | CPECU: 39, 11–12, 15, 17, 19, 20, 22, 24, 27, 29, 30, 32 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . |
| H_4 | CPECU: 33 | . | C | . | . | A | T | T | G | C | C | C | . | . | C | . | G | . | T | . | T | . | T | G | C | G |
| H_5 | CPECU: 23 | . | C | . | . | A | T | T | G | C | . | . | . | . | C | . | G | T | T | . | T | . | T | . | C | G |
| H_6 | CPECU: 28 | . | . | . | T | . | . | . | . | C | . | . | . | . | C | T | . | . | . | C | . | . | . | . | . | . |
| H_7 | CPECU: 2, 14 | . | C | . | . | A | T | T | G | C | C | C | A | C | C | . | G | . | T | . | T | . | T | G | C | G |
| H_8 | CPECU:1, 16, 18 | . | . | . | T | . | . | . | . | C | . | . | . | . | C | . | . | . | . | C | . | . | . | . | . | . |
| H_9 | CPECU: 10, 24 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | C | . | G | . | . | . | . |
| Population | Genetic Diversity Indices | Neutrality Test | ||||||
|---|---|---|---|---|---|---|---|---|
| N | Tnm | H | Hd | π | S | Tajima’s D (p) | Fu’s FS | |
| Pillareño Creole pig | 34 | 25 | 9 | 0.615 | 0.00968 | 25 | −0.44963 (0.454) | 1.794 (0.119) |
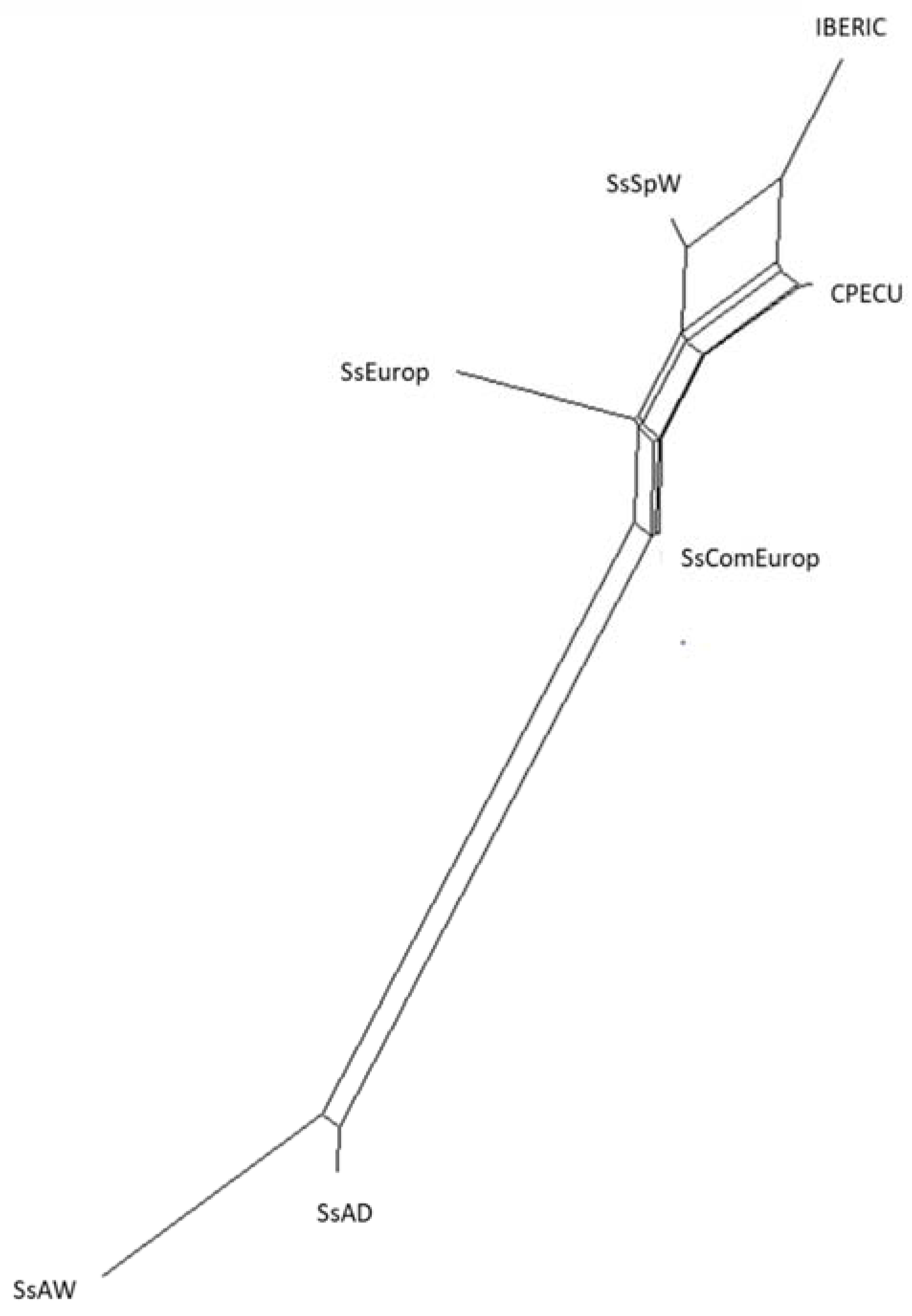
| Pig Population | CPECU | SSEUROP | IBERIC | SSCOMEUROP | SSAD | SSSPW | SSAW |
|---|---|---|---|---|---|---|---|
| CPECU | 0.00000 | ||||||
| SSEUROP | 0.19955 | 0.00000 | |||||
| IBERIC | 0.11760 | 0.25758 | 0.00000 | ||||
| SSCOMEUROP | 0.12242 | 0.14175 | 0.27297 | 0.00000 | |||
| SSAD | 0.47947 | 0.45415 | 0.56992 | 0.31056 | 0.00000 | ||
| SSSPW | 0.12439 | 0.19741 | 0.12655 | 0.13017 | 0.47835 | 0.00000 | |
| SSAW | 0.58502 | 0.55411 | 0.66240 | 0.42897 | 0.15217 | 0.56351 | 0.00000 |
| Pig Population | CPECU | SSEUROP | IBERIC | SSCOMEUROP | SSAD | SSSPW | SSAW |
|---|---|---|---|---|---|---|---|
| CPECU | 0 | ||||||
| SSEUROP | 0.22258 | 0 | |||||
| IBERIC | 0.12511 | 0.29784 | 0 | ||||
| SSCOMEUROP | 0.13059 | 0.15286 | 0.31879 | 0 | |||
| SSAD | 0.65291 | 0.60542 | 0.84378 | 0.37188 | 0 | ||
| SSSPW | 0.13284 | 0.21991 | 0.13531 | 0.13945 | 0.65075 | 0 | |
| SSAW | 0.87952 | 0.80769 | 1.08588 | 0.56031 | 0.16508 | 0.82900 | 0 |
| Source of Variation | Df | Sum of Squares | Variance Components | Percentage of Variation | Fixation Indices |
|---|---|---|---|---|---|
| Among groups | 3 | 47.108 | 0.06985 Va | 2.22 | |
| Among populations/within groups | 9 | 55.027 | 0.89171 Vb | 28.36 | FSC: 0.29004 FST: 0.30581 FCT: 0.02221 |
| Within populations | 84 | 183.350 | 2.18274 Vc | 69.42 | |
| Total | 96 | 811.794 | 3.14430 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vergara, A.M.C.; Martínez, A.M.; Bermejo, J.V.D.; Macri, M.; Nájera, P.R.A.; Duchi, N.A.D.; Vargas, P.A.T. A Matrilineal Study on the Origin and Genetic Relations of the Ecuadorian Pillareño Creole Pig Population through D-Loop Mitochondrial DNA Analysis. Animals 2021, 11, 3322. https://doi.org/10.3390/ani11113322
Vergara AMC, Martínez AM, Bermejo JVD, Macri M, Nájera PRA, Duchi NAD, Vargas PAT. A Matrilineal Study on the Origin and Genetic Relations of the Ecuadorian Pillareño Creole Pig Population through D-Loop Mitochondrial DNA Analysis. Animals. 2021; 11(11):3322. https://doi.org/10.3390/ani11113322
Chicago/Turabian StyleVergara, Amado Manuel Canales, Amparo Martínez Martínez, Juan Vicente Delgado Bermejo, Martina Macri, Pablo Rigoberto Andino Nájera, Nelson Antonio Duchi Duchi, and Paula Alexandra Toalombo Vargas. 2021. "A Matrilineal Study on the Origin and Genetic Relations of the Ecuadorian Pillareño Creole Pig Population through D-Loop Mitochondrial DNA Analysis" Animals 11, no. 11: 3322. https://doi.org/10.3390/ani11113322






