A Transcriptomic Analysis of Gonads from the Low-Temperature-Induced Masculinization of Takifugu rubripes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Consent to Participate
2.2. Fish Sample Collection
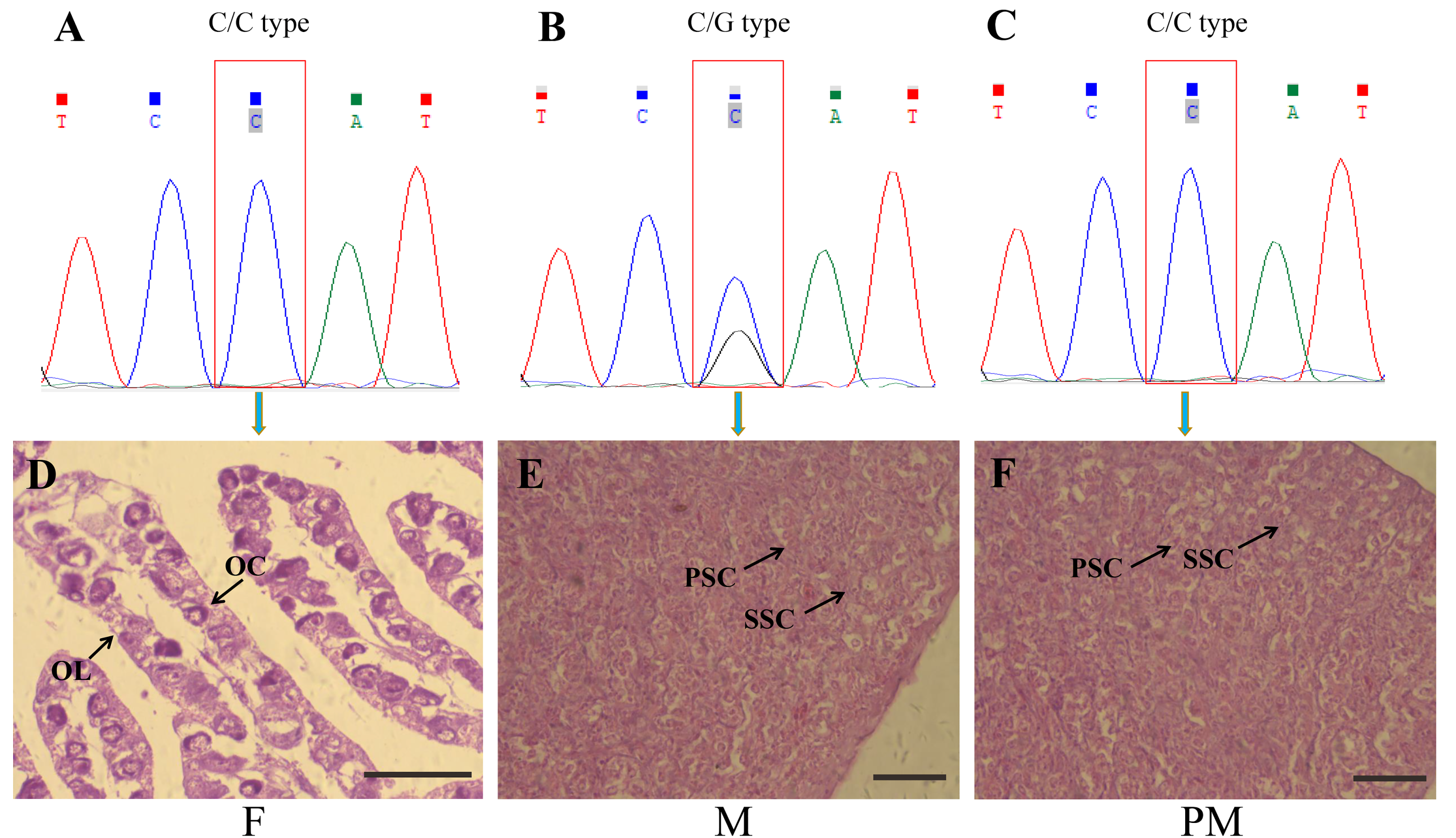
2.3. Screening of Pseudo-Males
2.4. RNA Collection and Sequencing
2.5. Processing of Raw Reads
2.6. Screening and Analysis of Differentially Expressed Genes (DEGs)
2.7. Quantitative PCR Verification
3. Results
3.1. Screening of Pseudo-Males
3.2. Data Statistics
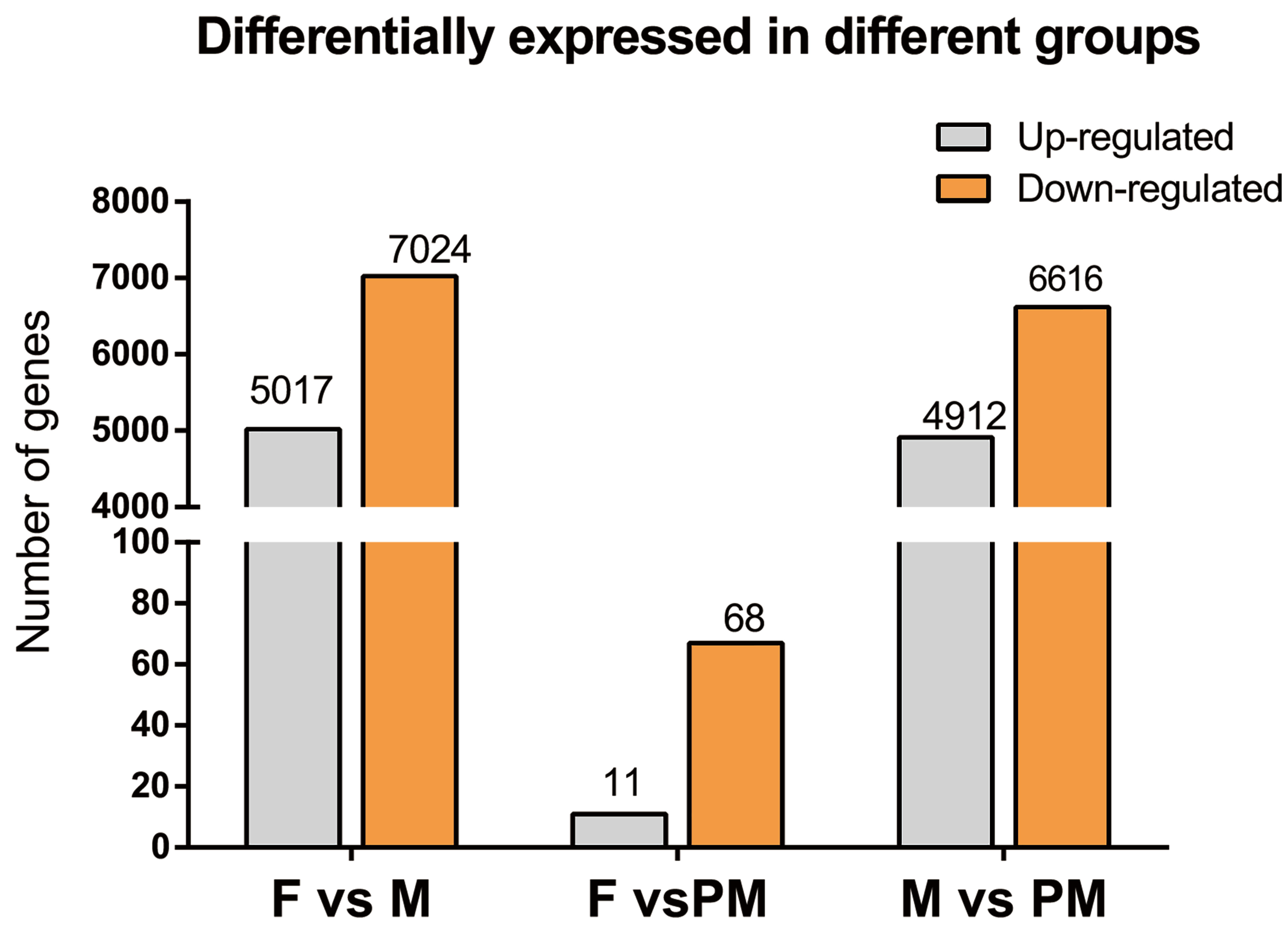
3.3. Sequencing Results and Interpretation of DEGs
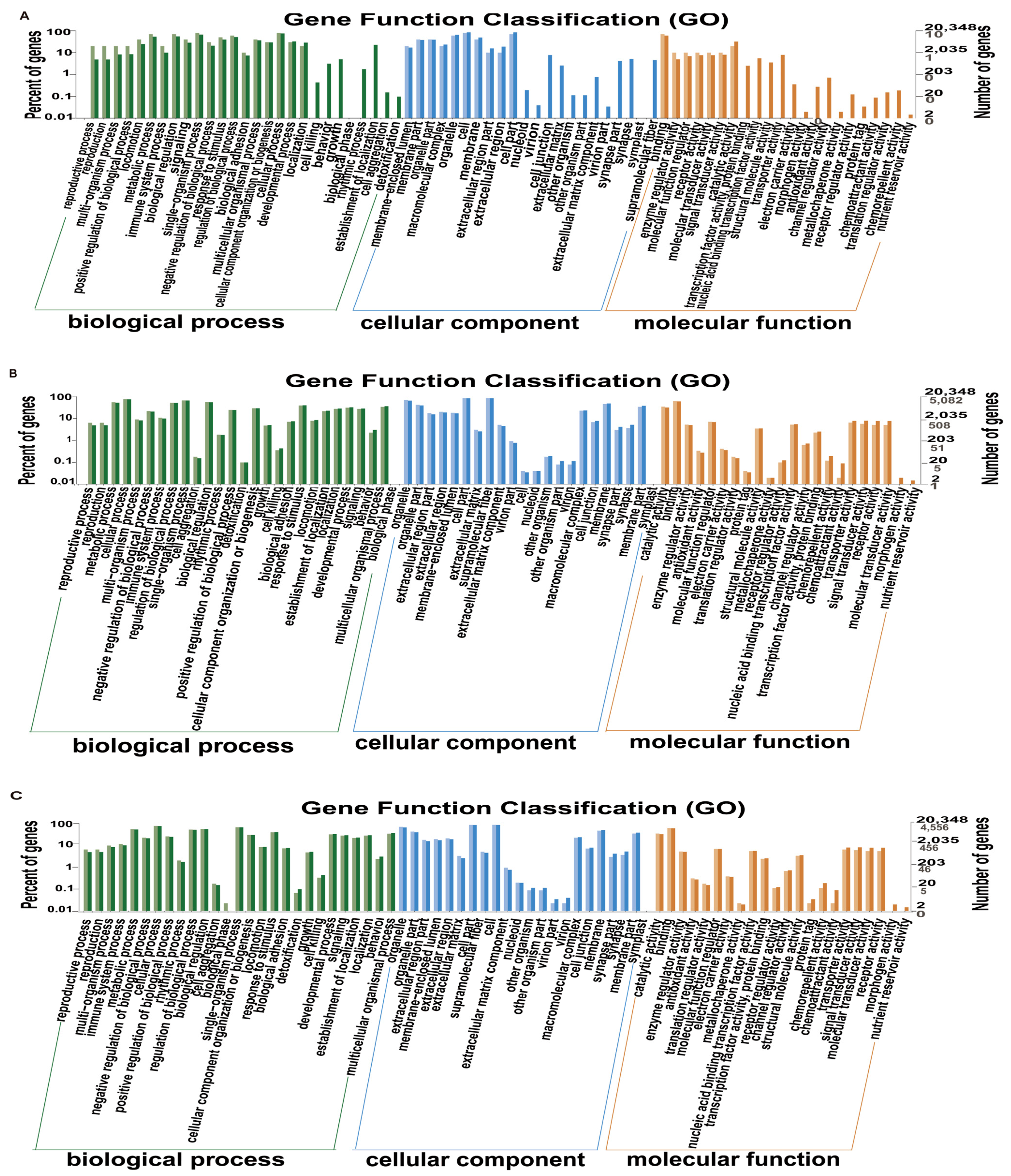
3.4. Gene Enrichment Analysis in Gene Ontology (GO)
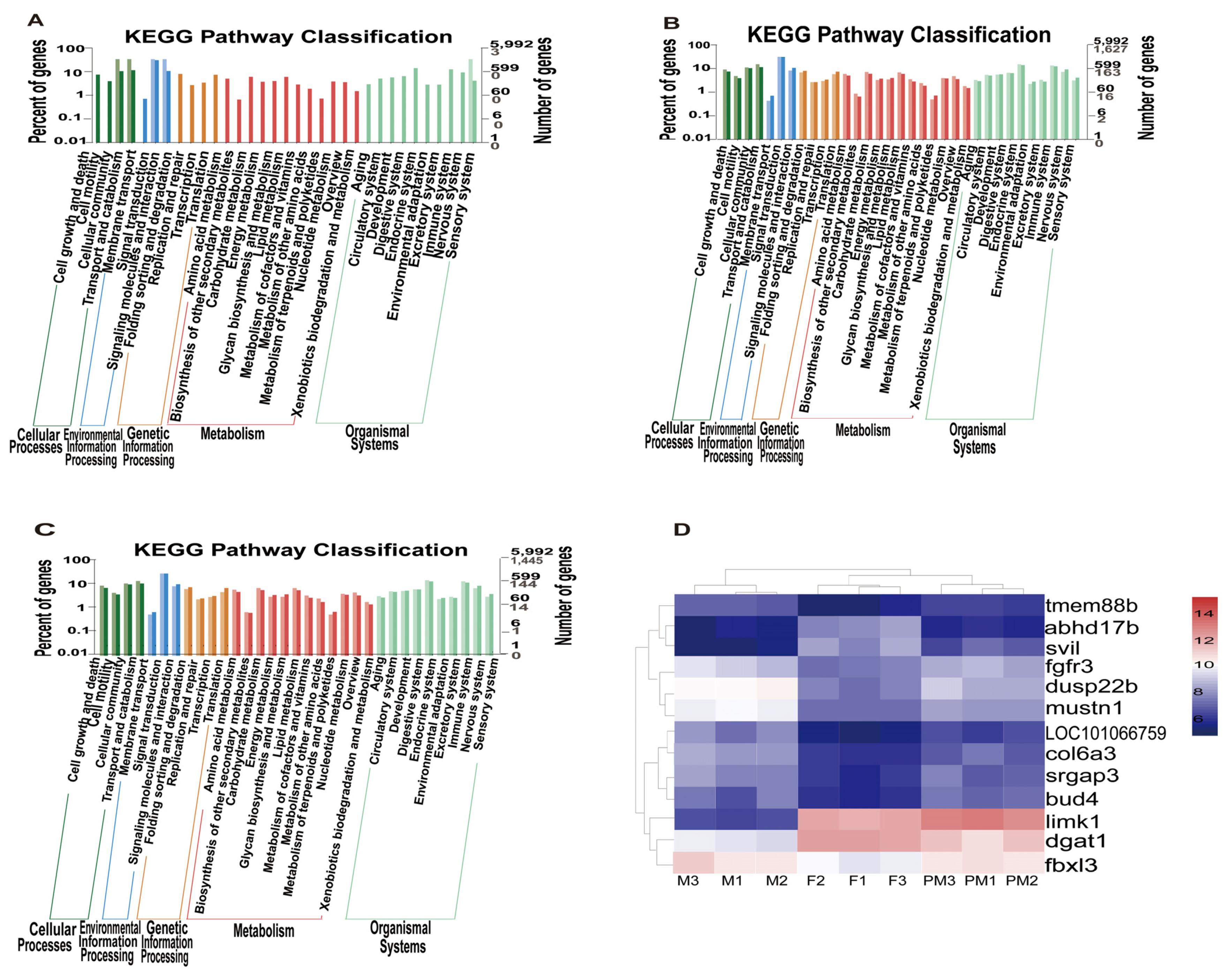
3.5. Gene Enrichment Analysis in the Kyoto Encyclopedia of Genes and Genomes (KEGG)
3.6. Clustering and Functional Analysis of DEGs
3.7. Analysis of Quantitative PCR Results
4. Discussion
| Gene ID | Annotation | Gene Name | Location in Chromosome |
|---|---|---|---|
| fbxl3 | F-box and leucine rich repeat protein 3 | fbxl3 | Chr 1:11,900,578–11,913,090 |
| LOC101075798 | collagen alpha-3(VI) chain-like | col6a3 | Chr 1:22,703,149–22,724,053 |
| LOC101063860 | fibroblast growth factor receptor 3-like | fgfr3 | Chr 2:10,771,162–10,816,015 |
| LOC105416485 | supervillin-like | svil | Chr 5:10,546,661–10,552,527 |
| LOC101061422 | LIM domain kinase 1-like | limk1 | Chr 11:3,287,016–3,309,899 |
| dgat1 | diacylglycerol O-acyltransferase 1 | dgat1 | Chr 12:9,699,595–9,709,017 |
| srgap3 | SLIT-ROBO Rho GTPase activating protein 3 | srgap3 | Chr 19:4,938,744–4,975,164 |
| tmem88b | transmembrane protein 88B | tmem88b | Chr 19:5,227,381–5,231,818 |
| LOC101077437 | bud site selection protein BUD4-like | bud4 | Chr 19:5,911,121–5,918,371 |
| mustn1 | musculoskeletal, embryonic nuclear protein 1 | mustn1 | Chr 19:4,520,735–4,521,811 |
| LOC101072874 | dual specificity protein phosphatase 22-B-like | dusp22b | Chr 22:145,042–148,673 |
| LOC101078060 | alpha/beta hydrolase domain-containing protein 17B | abhd17b | unknown |
| LOC101066759 | immune-type receptor | unknown |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, S. Sex Control and Cell Engineering Breeding of Fish; Science Press: Beijing, China, 2013. [Google Scholar]
- Ospina-Álvarez, N.; Piferrer, F. Temperature-dependent sex determination in fish revisited: Prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS ONE 2008, 3, e2837. [Google Scholar] [CrossRef] [Green Version]
- Díaz, N.; Piferrer, F. Lasting effects of early exposure to temperature on the gonadal transcriptome at the time of sex differentiation in the European sea bass, a fish with mixed genetic and environmental sex determination. BMC Genom. 2015, 16, 679. [Google Scholar] [CrossRef] [Green Version]
- Robledo, D.; Ribas, L.; Cal, R.; Sánchez, L.; Piferrer, F.; Martínez, P.; Viñas, A. Gene expression analysis at the onset of sex differentiation in turbot (Scophthalmus maximus). BMC Genom. 2015, 16, 973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Y.F.; Nan, P.; Zhang, W.; Wang, F.; Zhang, R.; Liang, T.; Ji, X.; Du, Q.; Chang, Z. Transcriptome analysis of three critical periods of ovarian development in Yellow River carp (Cyprinus carpio). Theriogenology 2018, 105, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, W.G.; He, M.X. Proteome and Transcriptome Analysis of Ovary, Intersex Gonads, and Testis Reveals Potential Key Sex Reversal/Differentiation Genes and Mechanism in Scallop Chlamys nobilis. Mar. Biotechnol. 2018, 20, 220–245. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Teng, J.; Zhao, Y.; Li, N.; Wang, H.; Ji, X. Gonad Transcriptome Analysis of High-Temperature-Treated Females and High-Temperature-Induced Sex-Reversed Neomales in Nile Tilapia. Int. J. Mol. Sci. 2018, 19, 689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Liu, X.; Jin, C.; Du, X.; He, Y.; Zhang, Q. Transcriptome Profiling Insights the Feature of Sex Reversal Induced by High Temperature in Tongue Sole Cynoglossus semilaevis. Front. Genet. 2019, 10, 522. [Google Scholar] [CrossRef] [Green Version]
- Lei, J. Japanese Pufferfish Culture Technology; Agriculture Press: Beijing, China, 2005; pp. 683–702. [Google Scholar]
- Li, J. Research on Masculinization of Takifugu Rubripes Induced by the Low Temperature. Master’s Thesis, Dalian Ocean University, Dalian, China, 2015; p. 45. [Google Scholar]
- Hattori, N.; Miyashita, S.; Sawada, Y. Effective masculinization method of tiger puffer by temperature control under culture condition. J. Fish. Soc. Japan 2012, 78, 87. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhou, Q.; Zhang, H.; Jiang, C.; Zhang, F. Effects of Temperature on Early Growth and Sex Differentiation of Takifugu rubripes. South China Fish. Sci. 2014, 10, 24–29. [Google Scholar] [CrossRef]
- Zhou, H.; Li, J.; Ma, H.; Li, Y.; Jiang, Z.; Li, X.; Xu, W.; Jiang, Z. Low-temperature-induced masculinization and histological observation of gonadal differentiation in Takifugu rubripes. J. Dalian Ocean Univ. 2015, 30, 41–47. [Google Scholar]
- Zhou, H.; Zhuang, Z.; Sun, Y.; Chen, Q.; Zheng, X.; Liang, Y.; Mahboob, S.; Wang, Q.; Zhang, R.; Al-Ghanim, K.A. Changes in DNA methylation during epigenetic associated sex reversal under low-temperature in Takifugu rubripes. PLoS ONE 2019, 14, e0221641. [Google Scholar] [CrossRef]
- Zhou, H.; Zhuang, Z.X.; Zhang, R.; Xu, Q.; Liang, Y.; Jiang, Z.; Li, X.; Ma, T.; Li, Y. Temperature-control-induced masculinization in tiger puffer Takifugu rubripes. J. Oceanol. Limnol. 2019, 37, 1125–1135. [Google Scholar] [CrossRef]
- Kamiya, T.; Kai, W.; Tasumi, S.; Oka, A.; Matsunaga, T.; Mizuno, N.; Fujita, M.; Suetake, H.; Suzuki, S.; Hosoya, S.; et al. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PloS Genet. 2012, 8, e1002798. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.; Salzberg, S. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Gui, J.F. Genetic Basis and Artificial Control of Sexuality and Production in Fish; Science Press: Beijing, China, 2007; p. 62. [Google Scholar]
- Yang, J.; Wu, H.D.; Fan, Z.T. Advances in Sex Determining Mechanisms in Fish. Fish. Econ. Res. 2007, 3, 14–19. [Google Scholar] [CrossRef]
- Wang, D.S.; Wu, T.L.; Zhang, Y.G. Advances in sex determination and its mechanisms in fish. J. Southwest China Norm. Univ. (Nat. Sci. Ed.) 2000, 25, 296–304. [Google Scholar] [CrossRef]
- Wang, Q.; Hao, X.; Liu, K.; Feng, B.; Li, S.; Zhang, Z.; Tang, L.; Mahboob, S.; Shao, C. Early response to heat stress in Chinese tongue sole (Cynoglossus semilaevis): Performance of different sexes, candidate genes and networks. BMC Genom. 2020, 21, 745. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, K.; Feng, B.; Zhang, Z.; Wang, R.; Tang, L.; Li, W.; Li, Q.; Piferrer, F.; Shao, C. Transcriptome of Gonads From High Temperature Induced Sex Reversal During Sex Determination and Differentiation in Chinese Tongue Sole, Cynoglossus semilaevis. Front. Genet. 2019, 10, 1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wang, Y.; Wang, S.; Liu, J.; Warren, W.; Mitreva, M.; Walter, R.B. Transcriptome analysis of female and male Xiphophorus maculatus Jp 163 A. PLoS ONE 2011, 6, e18379. [Google Scholar] [CrossRef]
- Yano, A.; Guyomard, R.; Nicol, B.; Jouanno, E.; Quillet, E.; Klopp, C.; Cabau, C.; Bouchez, O.; Fostier, A.; Guiguen, Y. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 2012, 22, 1423–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Liu, C.; Cao, X.; Huang, S.; Wang, W.; Wang, Y. Transcriptome Profile Analysis of Ovarian Tissues from Diploid and Tetraploid Loaches Misgurnus anguillicaudatus. Int. J. Mol. Sci. 2015, 16, 16017–16033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Xu, Q.; Zhang, R.; Zhuang, Z.; Ma, Y.; Wang, W.; Ma, T.; Sui, Y.; Liu, Y.; Cao, X. Gonadal transcriptome analysis of hybrid triploid loaches (Misgurnus anguillicaudatus)and their diploid and tetraploid parents. PLoS ONE 2018, 13, e0198179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Dsouza, N.R.; Zarate, Y.A.; Lombardo, R.; Hopkin, R.; Linehan, A.R.; Simpson, J.; McCarrier, J.; Agre, K.E.; Gavrilova, R.H.; et al. Genetic variants in dgat1 cause diverse clinical presentations of malnutrition through a specific molecular mechanism. Eur. J. Med. Genet. 2020, 63, 103817. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Wang, Y.; Cai, S.; Xu, M. dgat1 Expression Promotes Ovarian Cancer Progression and Is Associated with Poor Prognosis. J. Immunol. Res. 2021, 2021, 6636791. [Google Scholar] [CrossRef]
- Sakae, Y.; Oikawa, A.; Sugiura, Y.; Mita, M.; Nakamura, S.; Nishimura, T.; Suematsu, M.; Tanaka, M. Starvation causes female-to-male sex reversal through lipid metabolism in the teleost fish, medaka (Olyzias latipes). Biol. Open 2020, 9, bio050054. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Yao, Y.; Zhao, Y.; Liu, Y.; Liu, Z.; Hu, P.; Zhu, Z. Effects of PAK4/LIMK1/Cofilin-1 signaling pathway on proliferation, invasion, and migration of human osteosarcoma cells. J. Clin. Lab. Anal. 2020, 34, e23362. [Google Scholar] [CrossRef]
- Sebastian, S.; Rainer, D.; Steffen, J.; Spaich, S.; Kuhn, C.; Frank, D.; Berger, I.M.; Wiemann, S.; Korn, B.; Koegl, M.; et al. F-box and leucine-rich repeat protein 22 is a cardiac-enriched F-box protein that regulates sarcomeric protein turnover and is essential for maintenance of contractile function in vivo. Circ. Res. 2012, 111, 1504–1516. [Google Scholar] [CrossRef]
- Zhong, Y.; Xiang, G.; He, X.; Liu, S.; Zhang, X.; Zhang, J.; Chu, M.; Liu, Q. Preliminary Study on the Relationship between FBXL3 and FBXL21 Gene Expression and Seasonal Estrus in Sunite Sheep. Chin. J. Anim. Sci. 2020, 56, 40–44. [Google Scholar]
- Bonaldo, P.; Colombatti, A. The carboxyl terminus of the chicken alpha 3 chain of collagen VI is a unique mosaic structure with glycoprotein Ib-like, fibronectin type III, and Kunitz modules. J. Biol. Chem. 1989, 264, 20235–20239. [Google Scholar] [CrossRef]
- Sekine, Y.; Ikeda, O.; Hayakawa, Y.; Tsuji, S.; Imoto, S.; Aoki, N.; Sugiyama, K.; Matsuda, T. DUSP22/LMW-DSP2 regulates estrogen receptor a-mediated signaling through dephosphorylation of Ser-118. Oncogene 2007, 26, 6038–6049. [Google Scholar] [CrossRef] [Green Version]
- Lai, M.; Wang, C.; Yang, S.; Wu, C.; Sun, H.; Tsai, S.; Chuang, J.; Chen, Y.; Huang, B.M. The expression profiles of fibroblast growth factor 9 and its receptors in developing mice testes. Organogenesis 2016, 12, 61–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, Y.; Ma, Y.; Chen, K.; Mi, Y.; Fu, H.; Cui, D.; Jin, W. A link between the nuclear-localized srgap3 and the SWI/SNF chromatin remodeler Brg1. Mol Cell Neurosci. 2014, 60, 10–25. [Google Scholar] [CrossRef]
- Bacon, C.; Endris, V.; Rappold, G.A. The cellular function of srGAP3 and its role in neuronal morphogenesis. Mech. Dev. 2013, 130, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Abdelmoneim, A.; Abdu, A.; Chen, S.; Sepúlveda, M.S. Molecular signaling pathways elicited by 17α-ethinylestradiol in Japanese medaka male larvae undergoing gonadal differentiation. Aquat. Toxicol. 2019, 208, 187–195. [Google Scholar] [CrossRef]
- Clelland, E.S.; Kelly, S.P. Tight junction proteins in zebrafish ovarian follicles: Stage specific mRNA abundance and response to 17beta-estradiol, human chorionic gonadotropin, and maturation inducing hormone. Gen. Comp. Endocrinol. 2010, 168, 388–400. [Google Scholar] [CrossRef]
- Reyes, J.M.; Chitwood, J.L.; Ross, P.J. RNA-Seq profiling of single bovine oocyte transcript abundance and its modulation by cytoplasmic polyadenylation. Mol. Reprod. Dev. 2015, 82, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Pan, Z.; Zhu, C.; Chang, G.; Wu, N.; Ding, H.; Wang, H. Differential expression analysis and identification of sex-related genes by gonad transcriptome sequencing in estradiol-treated and non-treated Ussuri catfish Pseudobagrus ussuriensis. Fish Physiol. Biochem. 2021, 47, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ru, X.; Mustapha, U.F.; Jiang, D.; Huang, Y.; Pan, S.; Zhu, C.; Li, G. Characterization, expression, and regulatory effects of nr0b1a and nr0b1b in spotted scat (Scatophagus argus). Comp Biochem. Physiol B Biochem. Mol. Biol. 2021, 256, 110644. [Google Scholar] [CrossRef]

| Gene Name | Forward Primer (F)/Reverse Primer (R) (5′–3′) | |
|---|---|---|
| mustn1 | F | CGCCCAGAGGTAAGGAAAGA |
| R | TCTCTCGCATTCCTCCATTACT | |
| zglp1 | F | AGGAGTTCAGCAGAAACGGAG |
| R | GCAACGACAGGTTCGCATT | |
| sox3 | F | AACAACAGCAGCAACGAGGAT |
| R | CGTCGGTCAGAAGTTTCCAGT | |
| Sample | Raw Data | Q20 (%) | Q30 (%) | GC (%) | Clean Reads | Clean/Raw (%) |
|---|---|---|---|---|---|---|
| PM_1 | 49,139,712 | 97.10% | 92.89% | 51.59% | 45,934,400 | 93.48% |
| PM_2 | 50,438,140 | 97.02% | 92.71% | 51.81% | 47,102,818 | 93.39% |
| PM_3 | 50,378,422 | 97.36% | 93.44% | 50.58% | 47,490,250 | 94.27% |
| F_1 | 54,330,568 | 97.06% | 92.81% | 52.03% | 50,886,434 | 93.66% |
| F_2 | 53,706,244 | 97.05% | 92.80% | 52.53% | 50,073,694 | 93.24% |
| F_3 | 53,883,284 | 97.00% | 92.67% | 52.07% | 50,106,682 | 92.99% |
| M_1 | 59,716,736 | 97.01% | 92.92% | 48.48% | 55,127,742 | 92.32% |
| M_2 | 50,834,854 | 97.37% | 93.59% | 47.57% | 47,324,666 | 93.09% |
| M_3 | 45,212,258 | 97.25% | 93.35% | 47.08% | 42,104,402 | 93.13% |
| Sample | Total Reads | Total Mapped | Multiple Mapped | Uniquely Mapped |
|---|---|---|---|---|
| PM_1 | 45,934,400 (100.00%) | 41,885,381 (91.19%) | 3,352,916 (7.30%) | 38,532,465 (83.89%) |
| PM_2 | 47,102,818 (100.00%) | 43,676,065 (92.72%) | 3,219,480 (6.84%) | 40,456,585 (85.89%) |
| PM_3 | 47,490,250 (100.00%) | 43,686,271 (91.99%) | 3,200,997 (6.74%) | 40,485,274 (85.25%) |
| F_1 | 50,886,434 (100.00%) | 46,897,430 (92.16%) | 3,875,948 (7.62%) | 43,021,482 (84.54%) |
| F_2 | 50,073,694 (100.00%) | 46,745,301 (93.35%) | 3,348,249 (6.69%) | 43,397,052 (86.67%) |
| F_3 | 50,106,682 (100.00%) | 46,262,973 (92.33%) | 3,223,857 (6.43%) | 43,039,116 (85.89%) |
| M_1 | 55,127,742 (100.00%) | 50,553,238 (91.70%) | 2,220,041 (4.03%) | 48,333,197 (87.67%) |
| M_2 | 47,324,666 (100.00%) | 43,389,429 (91.68%) | 2,050,256 (4.33%) | 41,339,173 (87.35%) |
| M_3 | 42,104,402 (100.00%) | 38,849,353 (92.27%) | 1,651,395 (3.92%) | 37,197,958 (88.35%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Sun, Y.; Li, X.; Zhou, Z.; Ma, K.; Guo, W.; Liang, Y.; Xie, X.; Zhang, J.; Wang, Q.; et al. A Transcriptomic Analysis of Gonads from the Low-Temperature-Induced Masculinization of Takifugu rubripes. Animals 2021, 11, 3419. https://doi.org/10.3390/ani11123419
Zhou H, Sun Y, Li X, Zhou Z, Ma K, Guo W, Liang Y, Xie X, Zhang J, Wang Q, et al. A Transcriptomic Analysis of Gonads from the Low-Temperature-Induced Masculinization of Takifugu rubripes. Animals. 2021; 11(12):3419. https://doi.org/10.3390/ani11123419
Chicago/Turabian StyleZhou, He, Yuqing Sun, Xin Li, Ziyu Zhou, Kexin Ma, Wenxuan Guo, Yuting Liang, Xingyi Xie, Jingxian Zhang, Qian Wang, and et al. 2021. "A Transcriptomic Analysis of Gonads from the Low-Temperature-Induced Masculinization of Takifugu rubripes" Animals 11, no. 12: 3419. https://doi.org/10.3390/ani11123419






