Screening and Identification of Differential Ovarian Proteins before and after Induced Ovulation via Seminal Plasma in Bactrian Camels
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Handling Conditions
2.2. Semen Collection and SP Preparation
2.3. SP Injection and Sample Collection
2.4. Samples Preparation and Collection
2.5. iTRAQ Labeling and SCX Fractionation and LC-Electrospray Ionization MS/MS
2.6. Hormone Analysis
2.7. Western Blot
2.8. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.9. Immunofluorescence Staining
2.10. Data Analysis
3. Results
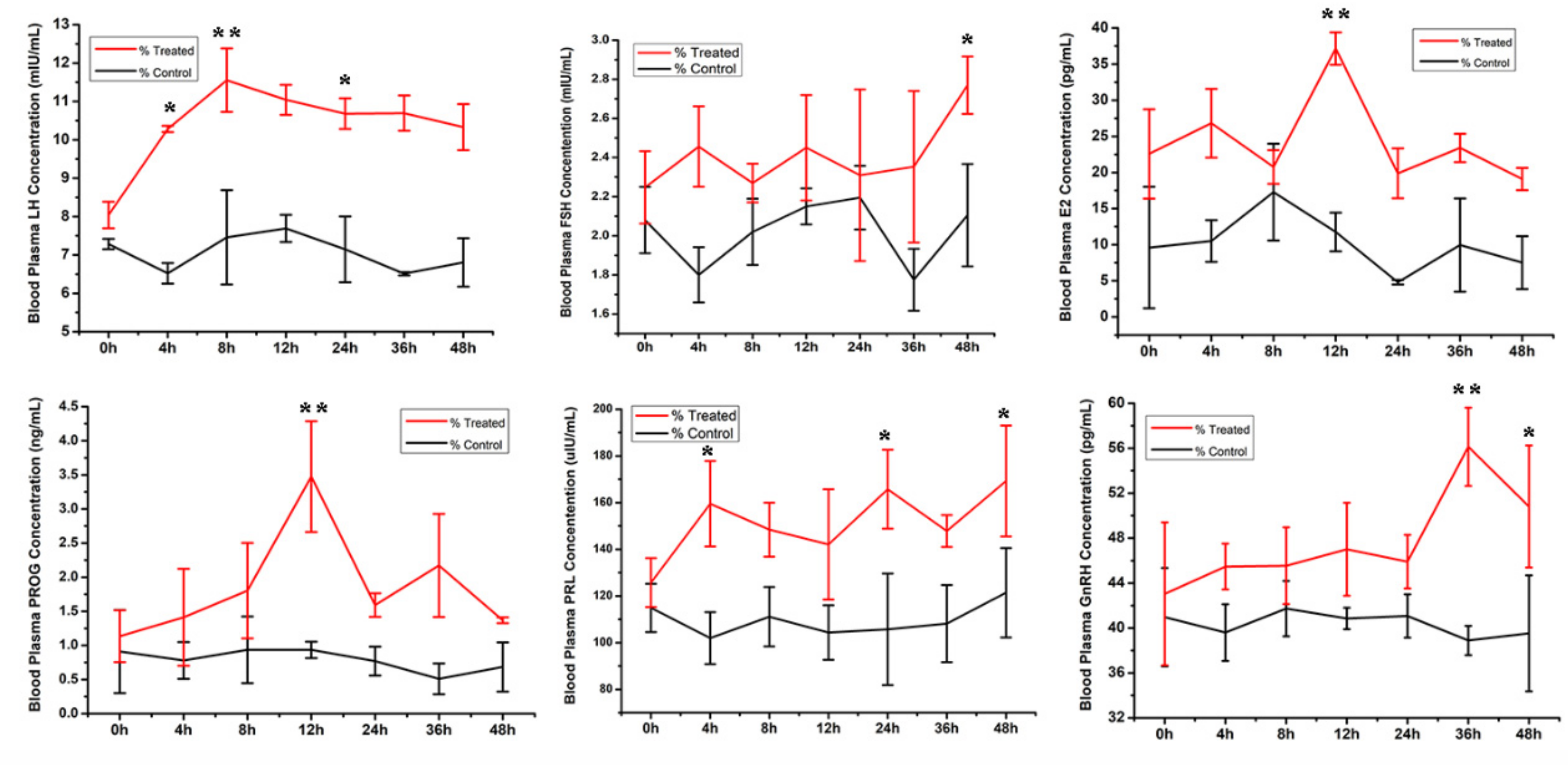
3.1. Follicle Size and Serum Hormone Levels
3.2. Protein Profiling and Identification of Differentially Expressed Proteins
3.3. Protein–Protein Interaction (PPI) Network Analysis
3.4. Expression Profile Verification
3.5. Analysis of Tissue Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Napoli, J.L. Functions of Intracellular Retinoid Binding-Proteins. Subcell. Biochem. 2016, 81, 21–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Ali, A.K.; Husayni, H.A.; Power, D.M. A comprehensive biochemical analysis of the blood of the camel (Camelus dromedarius). Comp. Biochem. Physiol. Part B Comp. Biochem. 1988, 89, 35–37. [Google Scholar] [CrossRef]
- Zhao, D.-B.; Bai, Y.-H.; Niu, Y.-W. Composition and characteristics of Chinese Bactrian camel milk. Small Rumin. Res. 2015, 127, 58–67. [Google Scholar] [CrossRef]
- Spies, H.G.; Pau, K.-Y.F.; Yang, S.-P. Coital and Estrogen Signals: A Contrast in the Preovulatory Neuroendocrine Networks of Rabbits and Rhesus Monkeys1. Biol. Reprod. 1997, 56, 310–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brüssow, K.-P.; Ratky, J.; Kanitz, W.; Becker, F. The Relationship between the Surge of LH Induced by Exogenous Gn-RH and the Duration of Ovulation in Gilts. Reprod. Domest. Anim. 1990, 25, 255–260. [Google Scholar] [CrossRef]
- Jöchle, W. Current research in coitus-induced ovulation: A review. J. Reprod. Fertil. Suppl. 1975, 22, 165–207. [Google Scholar]
- Fernandez-Baca, S.; Madden, D.H.L.; Novoa, C. Effect of different mating stimuli on induction of ovulation in the alpaca. J. Reprod. Fertil. 1970, 22, 261–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakker, J.; Baum, M.J. Neuroendocrine Regulation of GnRH Release in Induced Ovulators. Front. Neuroendocr. 2000, 21, 220–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratto, M.H.; Huanca, W.; Adams, G.P. Ovulation-inducing factor: A protein component of llama seminal plasma. Reprod. Biol. Endocrinol. 2010, 8, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.X.; Li, X.L.; Chen, B.X. Isolation of ovulation-inducing factors in the seminal plasma of Bactrian camel (Camelus bactrianus) by DEAE-cellulose chromatography. Reprod. Domest. Anim. 2001, 36, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.X.; Yuen, Z.X.; Pan, G.W. Semen-induced ovulation in the bactrian camel (Camelus bactrianus). Reproduction 1985, 74, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Ratto, M.H.; Huanca, W.; Singh, J.; Adams, G.P. Comparison of the effect of ovulation-inducing factor (OIF) in the seminal plasma of llamas, alpacas, and bulls. Theriogenology 2006, 66, 1102–1106. [Google Scholar] [CrossRef] [PubMed]
- Ratto, M.H.; Leduc, Y.A.; Valderrama, X.P.; van Straaten, K.E.; Delbaere, L.T.J.; Pierson, R.A.; Adams, G.P. The nerve of ovulation-inducing factor in semen. Proc. Natl. Acad. Sci. USA 2012, 109, 15042–15047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, M.E.; Smulders, J.P.; Guerra, M.; Valderrama, X.P.; Letelier, C.; Adams, G.P.; Ratto, M.H. Cetrorelix suppresses the preovulatory LH surge and ovulation induced by ovulation-inducing factor (OIF) present in llama seminal plasma. Reprod. Biol. Endocrinol. 2011, 9, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichtenwalner, A.B.; Woods, G.L.; Weber, J.A. Seminal collection, seminal characteristics and pattern of ejaculation in llamas. Theriogenology 1996, 46, 293–305. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Q.; Gan, Z.; Li, H.; Yang, Y.; Zhang, Y.; Zhao, X. Screening for reproductive biomarkers in Bactrian camel via iTRAQ analysis of proteomes. Reprod. Domest. Anim. 2019, 55, 189–199. [Google Scholar] [CrossRef]
- Greenwood, F.C.; Hunter, W.M.; Glover, J.S. The preparation of 131i-labelled human growth hormone of high specific radioactivity. Biochem. J. 1963, 89, 114–123. [Google Scholar] [CrossRef]
- Zhang, Q.; Gong, J.; Wang, X.; Wu, X.; Li, Y.; Ma, Y.; Zhang, Y.; Zhao, X. Molecular cloning, bioinformatics analysis and expression of insulin-like growth factor 2 from Tianzhu white yak, Bos grunniens. Int. J. Mol. Sci. 2014, 15, 504–524. [Google Scholar] [CrossRef] [Green Version]
- Otali, D.; Fredenburgh, J.; Oelschlager, D.K.; Grizzle, W.E. A standard tissue as a control for histochemical and immuno-histochemical staining. Biotech. Histochem. Off. Publ. Biol. Stain. Comm. 2016, 91, 309–326. [Google Scholar] [CrossRef] [Green Version]
- Berland, M.A.; Ulloa-Leal, C.; Barría, M.; Wright, H.; Dissen, G.A.; Silva, M.E.; Ojeda, S.R.; Ratto, M.H. Seminal Plasma Induces Ovulation in Llamas in the Absence of a Copulatory Stimulus: Role of Nerve Growth Factor as an Ovulation-Inducing Factor. Endocrinology 2016, 157, 3224–3232. [Google Scholar] [CrossRef] [PubMed]
- Adams, G.P.; Ratto, M.H.; Huanca, W.; Singh, J. Ovulation-Inducing Factor in the Seminal Plasma of Alpacas and Llamas1. Biol. Reprod. 2005, 73, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Adams, G.P.; Ratto, M.H.; Silva, M.E.; Carrasco, R.A. Ovulation-inducing factor (OIF/NGF) in seminal plasma: A review and update. Reproduction in Domestic Animals. 2016, 51, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.J.; Noh, Y.; Kim, M.S.; Jang, A.; Lee, C.E.; Myung, S.C. Steroidogenic effects of Taraxacum officinale extract on the levels of steroidogenic enzymes in mouse Leydig cells. Anim. Cells Syst. 2018, 22, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Ratto, M.H.; Delbaere, L.T.; Leduc, Y.A.; Pierson, R.A.; Adams, G.P. Biochemical isolation and purification of ovulation-inducing factor (OIF) in seminal plasma of llamas. Reprod. Biol. Endocrinol. 2011, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karsch, F.J. Central actions of ovarian steroids in the feedback regulation of pulsatile secretion of luteinizing hormone. Annu. Rev. Physiol. 1987, 49, 365–382. [Google Scholar] [CrossRef]
- Musa, B.; Abusineina, M. The oestrous cycle of the camel (Camelus dromedarius). Vet. Rec. 1978, 103, 556–557. [Google Scholar] [CrossRef] [PubMed]
- Sumar, J. Removal of the ovaries or ablation of the corpus luteum and its effect on the maintenance of gestation in the alpaca and llama. Acta Vet. Scand. Suppl. 1988, 83, 133–141. [Google Scholar]
- Zhao, X.; Chen, B. Ecophysiology and Reproduction of the Camelidae [M]; Gansu Science and Technology Press: Lanzhou, China, 1995. [Google Scholar]
- Silva, M.E.; Colazo, M.G.; Ratto, M.H. GnRH dose reduction decreases pituitary LH release and ovulatory response but does not affect corpus luteum (CL) development and function in llamas. Theriogenology 2012, 77, 1802–1810. [Google Scholar] [CrossRef]
- Paré, J.-F.; Roy, S.; Galarneau, L.; Bélanger, L. The Mouse Fetoprotein Transcription Factor (FTF) Gene Promoter Is Regulated by Three GATA Elements with Tandem E Box and Nkx Motifs, and FTF in Turn Activates the Hnf3β, Hnf4α, and Hnf1α Gene Promoters. J. Biol. Chem. 2001, 276, 13136–13144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lourenço, D.; Brauner, R.; Lin, L.; De Perdigo, A.; Weryha, G.; Muresan, M.; Boudjenah, R.; Guerra-Junior, G.; Maciel-Guerra, A.T.; Achermann, J.C.; et al. Mutations in NR5A1 associated with ovarian insufficiency. N. Engl. J. Med. 2009, 360, 1200–1210. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Choi, M.; Cavey, G.; Daugherty, J.; Suino, K.; Kovach, A.; Bingham, N.C.; Kliewer, S.A.; Xu, H.E. Crystallographic Identification and Functional Characterization of Phospholipids as Ligands for the Orphan Nuclear Receptor Steroidogenic Factor-1. Mol. Cell 2005, 17, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Achermann, J.C. Steroidogenic factor-1 (SF-1, Ad4BP, NR5A1) and disorders of testis development. Sex. Dev. 2008, 2, 200–209. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Ikeda, Y.; Parker, K.L. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 1994, 77, 481–490. [Google Scholar] [CrossRef]
- Ferraz-de-Souza, B.; Hudson-Davies, R.E.; Lin, L.; Parnaik, R.; Hubank, M.; Dattani, M.T.; Achermann, J.C. Sterol O-acyltransferase 1 (SOAT1, ACAT) is a novel target of steroidogenic factor-1 (SF-1, NR5A1, Ad4BP) in the human adrenal. J. Clin. Endocrinol. Metab. 2011, 96, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, T.; Minokoshi, Y. Systemic Glucoregulation by Glucose-Sensing Neurons in the Ventromedial Hypothalamic Nucleus (VMH). J. Endocr. Soc. 2017, 1, 449–459. [Google Scholar] [CrossRef] [Green Version]
- Ingraham, H.A.; Lala, D.S.; Ikeda, Y.; Luo, X.; Shen, W.H.; Nachtigal, M.W.; Abbud, R.; Nilson, J.H.; Parker, K.L. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994, 8, 2302–2312. [Google Scholar] [CrossRef] [Green Version]
- Mathews, L.S. Activin receptors and cellular signaling by the receptor serine kinase family. Endocr. Rev. 1994, 15, 310–325. [Google Scholar] [CrossRef]
- Bondestam, J.; Horelli-Kuitunen, N.; Hildén, K.; Ritvos, O.; Aaltonen, J. Assignment of ACVR2 and ACVR2B the human activin receptor type II and IIB genes to chromosome bands 2q22.2-->q23.3 and 3p22 and the human follistatin gene (FST) to chromosome 5q11.2 by FISH. Cytogenet. Cell Genet. 1999, 87, 219–220. [Google Scholar] [CrossRef]
- Gospodarowicz, D.; Lau, K. Pituitary follicular cells secrete both vascular endothelial growth factor and follistatin. Biochem. Biophys. Res. Commun. 1989, 165, 292–298. [Google Scholar] [CrossRef]
- Shimasaki, S.; Koga, M.; Buscaglia, M.L.; Simmons, D.M.; Bicsak, T.A.; Ling, N. Follistatin Gene Expression in the Ovary and Extragonadal Tissues. Mol. Endocrinol. 1989, 3, 651–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, H.H.-C. The pathway to femaleness: Current knowledge on embryonic development of the ovary. Mol. Cell. Endocrinol. 2005, 230, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Carrasco, R.; Singh, J.; Adams, G.P. The dynamics of trkA expression in the bovine ovary are associated with a luteotrophic effect of ovulation-inducing factor/nerve growth factor (OIF/NGF). Reprod. Biol. Endocrinol. 2016, 14, 47. [Google Scholar] [CrossRef] [Green Version]
- Griffond, B.; Deray, A.; Fellmann, D.; Ciofi, P.; Croix, D.; Bugnon, C. Colocalization of prolactin- and dynorphin-like sub-stances in a neuronal population of the rat lateral hypothalamus. Neurosci. Lett. 1993, 156, 91–95. [Google Scholar] [CrossRef]
- Freeman, M.E.; Kanyicska, B.; Lerant, A.; Nagy, G. Prolactin: Structure, Function, and Regulation of Secretion. Physiol. Rev. 2000, 80, 1523–1631. [Google Scholar] [CrossRef] [PubMed]
- DeVito, W.J.; Stone, S.; Avakian, C. Stimulation of Hypothalamic Prolactin Release by Veratridine and Angiotensin II in the Female Rat: Effect of Ovariectomy and Estradiol Administration. Neuroendocrinology 1991, 54, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Daza, D.O.; Larhammar, D. Evolution of the growth hormone, prolactin, prolactin 2 and somatolactin family. Gen. Comp. Endocrinol. 2018, 264, 94–112. [Google Scholar] [CrossRef] [PubMed]
- Lamberts, S.W.J.; De Quijada, M.; Klijn, J.G.M. The effect of tamoxifen on GH and PRL secretion by human pituitary tumors. J. Endocrinol. Investig. 1980, 3, 343–347. [Google Scholar] [CrossRef]
- Mednick, D.L.; Barkley, M.S.; Geschwind, I. Regulation of progesterone secretion by LH and prolactin during the first half of pregnancy in the mouse. J. Reprod. Fertil. 1980, 60, 201–207. [Google Scholar] [CrossRef] [Green Version]
- Gibori, G.; Richards, J.S. Dissociation of Two Distinct Luteotropic Effects of Prolactin: Regulation of Luteinizing Hormone-Receptor Content and Progesterone Secretion during Pregnancy. Endocrinology 1978, 102, 767–774. [Google Scholar] [CrossRef] [PubMed]





| Primer | Sequence 5′–3′ | Product Length (bp) | Accession No. |
|---|---|---|---|
| GAPDH | gtggagccaagagggtcat | 232 | NW_011514262.1 |
| gggccatccacagtcttct | |||
| PRL | gggcagagggttcatgact | 176 | XM_010974249.1 |
| taccccgcacttctgtgac | |||
| NR5A1 | tggtgttcgaccacatctacc | 221 | XM_010956204.1 |
| gaggctgaagaggatgaggaa | |||
| FST | agacctgtcgggatgttttct | 183 | XM_010966300.1 |
| ggaggcaggtagcctttctta |
| Different Expression | Protein Name |
|---|---|
| UP | CRT, GABRB2, HDGF, CLIC1, LOC10507404, PURH, PPP1CB, CLIC2, STX11, SAP30BP, SIPA1L1, PIP4K2C, ABCB9, CD63, TTC19, SOX1, EXOSC8, SAT2, CTR9, TMSB10, LDLR, F13A1, P3H4, LOC105068711, FGGY, TBC1D24, TMEM87A, Slc22A17, HSL, FOXK1, HSPB6, REXO2, Ighg1a, SEBOX, ATP1B1, MRPS26, CASK, Mrpl24, CCDC43, PHC2, UBE2N, MFF, LRRFIP1, NRIP2, COPE, SMARCB1, KRT8, CDHR2, C17ORF67, AGPS, HDAC3, CAMKK1, PON1, CTSS, GPX1, FKBP14, MRPL14, MTHFD2, CAPN2, THBS3, DDX19A, LGALSL, P3H1, JUND, BORCS6, SERPINH1, PTGES3, PTMS, TFCP2, TNFaIP2, NFYB, WAC, PTPN6, LAMTOR1, CDH1, LIPG, SPARC, BTF3, STMN1, MANF, UFM1, Igh, PRL, REX1BD, CNN3, NUDC, CNPY3, MBP, EDEM1, CHST14, PLCH1, PYCR1, STX2, SEC23B, CDC42EP5, GBP1, KRT39, LOC105082461, CRMP1, PCNP, CIAPIN1, HDDC3, B2M, MFSD10, PPL, GJA1, RARRES2, TFAP2B, SYVN1, FRAS1, MED1, PLP1, SMAP2, PSAT1, ELOB, QPCT, ATL2, SLC25A46, SCCA, GLT8D2, PSTPIP2, AGG, TGFB2, NDEL1, TCHH, BZW2, VPS13D, GSDMD, CRYZL1, CRABP2, CSTB, BCKDHB, RAB43, SPATA5, eEF2K, PRRC1, RER1, PSMF1, MRPL50, GABARAPL2, CTRL, SLC13A5, RABL6, MTRR, MRC2, NPC1, PLIN3, KRT7, PSME2, NAT10, COL6A5, CSN2, NSD1, SYT1, FKBP10, FDX1, TXN2, LGALS3, SLC35A3, HCLS1, MCRIP1, LOC105069208, SREK1, GLS, RPE, PPA1, LOC105070504, LRRC41, OTUB1, TSFM, TEAD1, LOC105073173, CDC123, IRF3, CALU, FABP5, KCNAB2, RWDD1, P4HA2, LOC105067606, SAR1B, MRPS25, LOC105069779, ERAP2, LOC105066647, LOC105067240, LBP, SDC4, TCN1, COPZ1, SAR1A, SLC46A1, JPT2, KRT18, VSNL1, DDX59, LOC105079327, CD109, CNN2, MAN2A1, ICOSLG, LOC105077404, PPP1R2, SLC1A5, S100A6, SEC11C, CTSV, RCN3, UBR2, PSMB10, TPT1, LOC105075883, IRF2BP2, LOC105074738, ARMCX3, ESPL1, HP, C1QTNF3, BABAM1, ELOC, GOLIM4, MRTFB, CDV3, ARHGAP32, TMEM94, TP53I3, GFPT2, CEP131, TYMS, PLPP3, PAK1, PPCA, LETMD1, GFAP, LRRC8D, MCF2L, ITIH4, SIGLEC1, SEC13, EI24, P3H3, CHMP5, POLR2I, WARS1, SELENOH, aLG11, ITIH3, MRFAP1, CEP104, MRPS31, DHCR24, MRPS36, PLA2G6, VTA1, HYPK, STAG2. |
| Down | ELMO2, LAP2, NAT14, ESPN, LPAAT1, DT S7, NEFL, Wnt2b, MCAM, Rint1, ARF5, BRPF1, WASH1, c-Rel, TPD52, KRT19, TSTD3, CDK19, PTPP, EXOC6, CRT11, TMEM209, LOC105080173, ISPS, Di-Ras1, RIM11, FST, GGTB, P400, TESTIN, PRELP, LMOD1, TINAGL1, TNFAIP8L3, HOXD8, SCO2, ZCCHC3, Septin4, NIPSNAP1, NUMB, ZBED5, DHX35, PATZ, ALDH3B1, LAMA4, RPL11, NR5A1, PDX1, CLYBL, MYH11, ABCC8, VPS4A, GLRX1, LIMS2, PPP1R10, LOC105068711, KRT1, Ybx3, MVI, ANAPC7, LAMB2, MYZAP, MED1, ZNF410, PFDN6, Smoothelin, OR, PYCR2, MSH3, PNPLA7, CAMK2G, KRT17, PPBP, DDX31, MRPL19, ZER1, MSTO1, KCR1, RPS27L, RyR3, QXPO4, Desmin, FBXO7, HSPA2, PGPEP1, MYO7, Lamc1, A4D1P6, NOB1, ZNF326, TTC21B, CBL, RMC1, TRIM32, LIN9, SHPK, EPC1, PBX2, SVEP1, DCTN4, PRKCD, HEBP2, EML6, DDT, LENG8, RPRD2, ECHDC3, PLIN4, H2A, BAG2, FLNA, HMCN2, PLEKHH2, OSTC, CAVIN3, AHSP, GTPBP3, ARID4B, ART4, TINF2, CTU2, MEN1, RNF123, MARK1, CCDC127. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhang, Q.; Li, Y.; Zhao, X.; Zhang, Y. Screening and Identification of Differential Ovarian Proteins before and after Induced Ovulation via Seminal Plasma in Bactrian Camels. Animals 2021, 11, 3512. https://doi.org/10.3390/ani11123512
Wang Q, Zhang Q, Li Y, Zhao X, Zhang Y. Screening and Identification of Differential Ovarian Proteins before and after Induced Ovulation via Seminal Plasma in Bactrian Camels. Animals. 2021; 11(12):3512. https://doi.org/10.3390/ani11123512
Chicago/Turabian StyleWang, Qi, Quanwei Zhang, Yina Li, Xingxu Zhao, and Yong Zhang. 2021. "Screening and Identification of Differential Ovarian Proteins before and after Induced Ovulation via Seminal Plasma in Bactrian Camels" Animals 11, no. 12: 3512. https://doi.org/10.3390/ani11123512
APA StyleWang, Q., Zhang, Q., Li, Y., Zhao, X., & Zhang, Y. (2021). Screening and Identification of Differential Ovarian Proteins before and after Induced Ovulation via Seminal Plasma in Bactrian Camels. Animals, 11(12), 3512. https://doi.org/10.3390/ani11123512






