Role of JAK-STAT Pathway in Broiler Chicks Fed with Chestnut Tannins
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals, Housing, and Treatments
2.2. Sample Collection and Processing
2.3. Bacterial Detection
2.4. Chicken-Specific Kinome (Peptide) Array
2.5. Real-Time Quantitative RT-PCR Assay
2.6. Data Analysis: Kinome Array
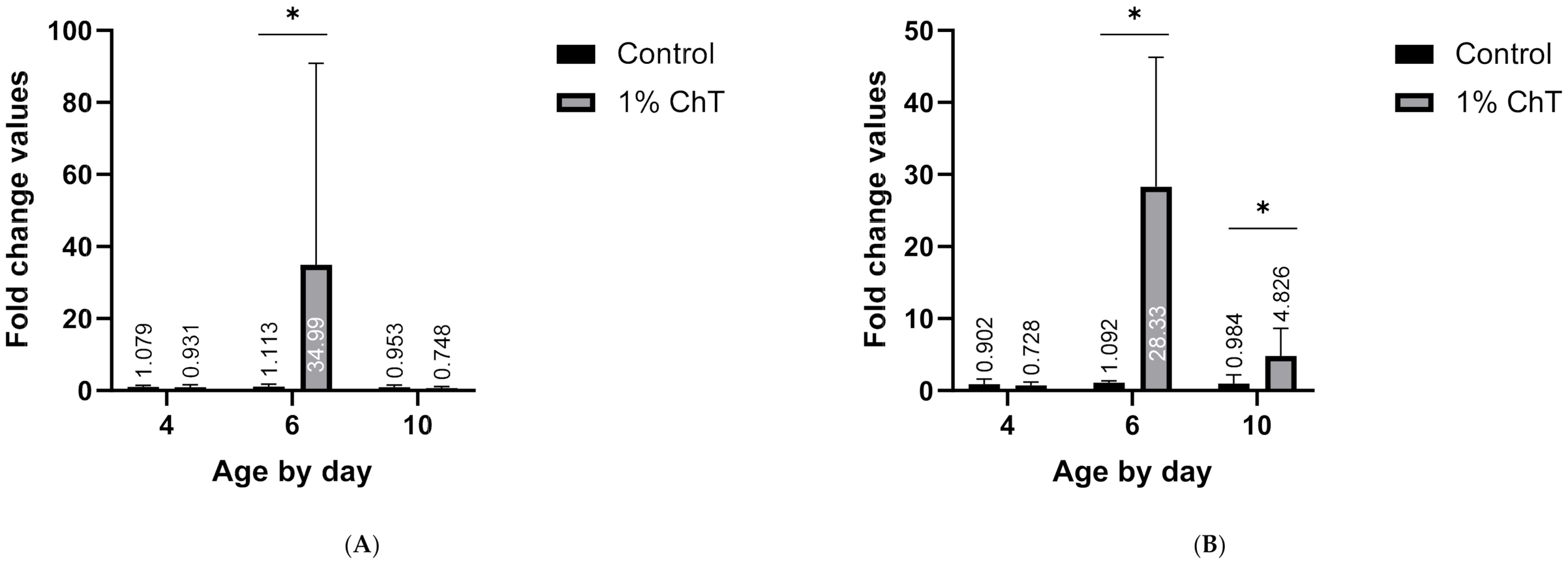
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Statement
References
- Lillehoj, H.; Liu, Y.; Calsamiglia, S.; Fernandez-Miyakawa, M.E.; Chi, F.; Cravens, R.L.; Oh, S.; Gay, C.G. Phytochemicals as antibiotic alternatives to promote growth and enhance host health. Vet. Res. 2018, 49, 1. [Google Scholar] [CrossRef]
- Ren, H.; Vahjen, W.; Dadi, T.; Saliu, E.-M.; Boroojeni, F.G.; Zentek, J. Synergistic effects of probiotics and phytobiotics on the intestinal microbiota in young broiler chicken. Microorganisms 2019, 7, 684. [Google Scholar] [CrossRef]
- Diaz Carrasco, J.M.; Redondo, L.M.; Redondo, E.A.; Dominguez, J.E.; Chacana, A.P.; Fernandez Miyakawa, M.E. Use of plant extracts as an effective manner to control Clostridium perfringens induced necrotic enteritis in poultry. BioMed. Res. Int. 2016, 2016, 3278359. [Google Scholar] [CrossRef]
- Díaz Carrasco, J.M.; Redondo, E.A.; Pin Viso, N.D.; Redondo, L.M.; Farber, M.D.; Fernández Miyakawa, M.E. Tannins and bacitracin differentially modulate gut microbiota of broiler chickens. BioMed. Res. Int. 2018, 2018, 1879168. [Google Scholar] [CrossRef]
- Huang, Q.; Liu, X.; Zhao, G.; Hu, T.; Wang, Y. Potential and challenges of tannins as an alternative to in-feed antibiotics for farm animal production. Anim. Nutr. 2018, 4, 137–150. [Google Scholar] [CrossRef]
- Brus, M.; Gradišnik, L.; Trapečar, M.; Škorjanc, D.; Frangež, R. Beneficial effects of water-soluble chestnut (Castanea sativa mill.) tannin extract on chicken small intestinal epithelial cell culture. Poult. Sci. 2018, 97, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Molino, S.; Casanova, N.A.; Rufián Henares, J.Á.; Fernandez Miyakawa, M.E. Natural tannin wood extracts as a potential food ingredient in the food industry. J. Agric. Food Chem. 2020, 68, 2836–2848. [Google Scholar] [CrossRef]
- Liu, H.W.; Li, K.; Zhao, J.S.; Deng, W. Effects of chestnut tannins on intestinal morphology, barrier function, pro-inflammatory cytokine expression, microflora and antioxidant capacity in heat-stressed broilers. J. Anim. Physiol. Anim. Nutr. 2018, 102, 717–726. [Google Scholar] [CrossRef]
- Song, P.; Zhang, R.; Wang, X.; He, P.; Tan, L.; Ma, X. Dietary grape-seed procyanidins decreased postweaning diarrhea by modulating intestinal permeability and suppressing oxidative stress in rats. J. Agric. Food Chem. 2011, 59, 6227–6232. [Google Scholar] [CrossRef]
- Williams, A.R.; Andersen-Civil, A.I.S.; Zhu, L.; Blanchard, A. Dietary phytonutrients and animal health: Regulation of immune function during gastrointestinal infections. J. Anim. Sci. 2020, 98, 1–11. [Google Scholar] [CrossRef]
- Manning, G. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Arsenault, R.; Griebel, P.; Napper, S. Peptide arrays for kinome analysis: New opportunities and remaining challenges. Proteomics 2011, 11, 4595–4609. [Google Scholar] [CrossRef] [PubMed]
- Jalal, S.; Arsenault, R.; Potter, A.A.; Babiuk, L.A.; Griebel, P.J.; Napper, S. Genome to kinome: Species-specific peptide arrays for kinome analysis. Sci. Signal. 2009, 2, pl1. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Cardoso Dal Pont, G.; Farnell, M.B.; Jarvis, S.; Battaglia, M.; Arsenault, R.J.; Kogut, M.H. Supplementing chestnut tannins in the broiler diet mediates a metabolic phenotype of the ceca. Poult. Sci. 2020, 100, 47–54. [Google Scholar] [CrossRef]
- Arsenault, R.J.; Lee, J.T.; Latham, R.; Carter, B.; Kogut, M.H. Changes in immune and metabolic gut response in broilers fed β-mannanase in β-mannan-containing diets. Poult. Sci. 2017, 96, 4307–4316. [Google Scholar] [CrossRef]
- Eldaghayes, I.; Rothwell, L.; Williams, A.; Withers, D.; Balu, S.; Davison, F.; Kaiser, P. Infectious bursal disease virus: Strains that differ in virulence differentially modulate the innate immune response to infection in the chicken bursa. Viral Immunol. 2006, 19, 83–91. [Google Scholar] [CrossRef]
- Kogut, M.H.; Rothwell, L.; Kaiser, P. Differential regulation of cytokine gene expression by avian heterophils during receptor-mediated phagocytosis of opsonized and nonopsonized Salmonella enteritidis. J. Interferon Cytokine Res. 2003, 23, 319–327. [Google Scholar] [CrossRef]
- Kaiser, P.; Wigley, P.; Burnside, J.; Barrow, P.A.; Galyov, E.E.; Rothwell, L. Differential cytokine expression in avian cells in response to invasion by Salmonella typhimurium, Salmonella enteritidis and Salmonella gallinarum. Microbiology 2000, 146, 3217–3226. [Google Scholar] [CrossRef]
- Trost, B.; Kindrachuk, J.; Määttänen, P.; Napper, S.; Kusalik, A. PIIKA 2: An expanded, web-based platform for analysis of kinome microarray data. PLoS ONE 2013, 8, e80837. [Google Scholar] [CrossRef]
- Li, Y.; Arsenault, R.J.; Trost, B.; Slind, J.; Griebel, P.J.; Napper, S.; Kusalik, A. A systematic approach for analysis of peptide array kinome data. Sci. Signal. 2012, 5, pl2. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Kogut, M.H.; Yin, X.; Yuan, J.; Broom, L. Gut health in poultry. CAB Rev. 2017, 12, 1–7. [Google Scholar] [CrossRef]
- Broom, L.J.; Kogut, M.H. Deciphering desirable immune responses from disease models with resistant and susceptible chickens. Poult. Sci. 2019, 98, 1634–1642. [Google Scholar] [CrossRef] [PubMed]
- Kogut, M.H. The effect of microbiome modulation on the intestinal health of poultry. Anim. Feed Sci. Tech. 2019, 250, 32–40. [Google Scholar] [CrossRef]
- Bartunek, P.; Koritschoner, N.P.; Brett, D.; Zenke, M. Molecular cloning, expression and evolutionary analysis of the avian tyrosine kinase JAK1. Gene 1999, 230, 129–136. [Google Scholar] [CrossRef]
- Truong, A.D.; Rengaraj, D.; Hong, Y.; Hoang, C.T.; Hong, Y.H.; Lillehoj, H.S. Analysis of JAK-STAT signaling pathway genes and their microRNAs in the intestinal mucosa of genetically disparate chicken lines induced with necrotic enteritis. Vet. Immunol. Immunopathol. 2017, 187, 1–9. [Google Scholar] [CrossRef]
- Huang, Y.; Wange, R.L. T cell receptor signaling: Beyond complex complexes. J. Biol. Chem. 2004, 279, 28827–28830. [Google Scholar] [CrossRef]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018, 27, 1984–2009. [Google Scholar]
- Zhang, Y.; Zhang, L.; Zuo, Q.; Wang, Y.; Zhang, Y.; Xu, Q.; Li, B.; Chen, G. JAK-STAT signaling regulation of chicken embryonic stem cell differentiation into male germ cells. In Vitro Cell. Dev. Biol. 2017, 53, 728–743. [Google Scholar] [CrossRef]
- Cousins, E.; Gao, Y.; Sandford, G.; Nicholas, J. Human herpesvirus 8 viral interleukin-6 signaling through gp130 promotes virus replication in primary effusion lymphoma and endothelial cells. J. Virol. 2014, 88, 12167–12172. [Google Scholar]
- Stross, C.; Radtke, S.; Clahsen, T.; Gerlach, C.; Volkmer-Engert, R.; Schaper, F.; Heinrich, P.C.; Hermanns, H.M. Oncostatin M receptor-mediated signal transduction is negatively regulated by SOCS3 through a receptor tyrosine-independent mechanism. J. Biol. Chem. 2006, 281, 8458–8468. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, J.S. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- Seif, F.; Khoshmirsafa, M.; Aazami, H.; Mohsenzadegan, M.; Sedighi, G.; Bahar, M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun. Signal. 2017, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef] [PubMed]
| Immune Pathways | ||||||
|---|---|---|---|---|---|---|
| Day 4 | Day 6 | Day 10 | ||||
| Identified Pathways | Number of Peptides | p-Value | Number of Peptides | p-Value | Number of Peptides | p-Value |
| Chemokine signaling pathway | 23 | 9.19 × 10−18 | 26 | 9.19 × 10−19 | 18 | 7.10 × 10−13 |
| T cell receptor signaling pathway | 14 | 7.55 × 10−12 | 25 | 5.85 × 10−23 | 12 | 7.50 × 10−10 |
| Jak-STAT signaling pathway | 13 | 1.38 × 10−8 | 19 | 7.82 × 10−13 | 15 | 1.31 × 10−10 |
| B cell receptor signaling pathway | 13 | 2.94 × 10−12 | 19 | 3.27 × 10−18 | 12 | 3.13 × 10−11 |
| Fc gamma R-mediated phagocytosis | 13 | 2.81 × 10−11 | 13 | 4.06 × 10−10 | - | - |
| TNF signaling pathway | 13 | 2.21 × 10−10 | 21 | 1.17 × 10−17 | 10 | 1.72 × 10−7 |
| Wnt signaling pathway | 13 | 4.24 × 10−9 | 11 | 3.30 × 10−6 | - | - |
| Apoptosis | 12 | 2.09 × 10−8 | 19 | 5.79 × 10−14 | - | - |
| Toll-like receptor signaling pathway | 11 | 1.46 × 10−8 | 20 | 6.03 × 10−17 | 13 | 9.43 × 10−11 |
| Natural killer cell mediated cytotoxicity | 11 | 8.57 × 10−8 | 16 | 1.76 × 10−11 | - | - |
| NOD-like receptor signaling pathway | 10 | 8.08 × 10−6 | 18 | 1.22 × 10−11 | 12 | 1.11 × 10−7 |
| Th17 cell differentiation | 10 | 1.43 × 10−7 | 22 | 3.36 × 10−19 | 10 | 1.11 × 10−7 |
| IL-17 signaling pathway | - | - | 16 | 3.62 × 10−13 | - | - |
| Inflammatory mediator regulation of TRP channels | - | - | 10 | 6.08 × 10−7 | - | - |
| JAK-STAT Signaling Pathway | ||||||||
|---|---|---|---|---|---|---|---|---|
| Days of Necropsy | ||||||||
| 4 | 6 | 10 | ||||||
| Proteins | UniProt ID | p-Site | Fold Change | p-Value | Fold Change | p-Value | Fold Change | p-Value |
| AKT3 | Q9Y243 | S476/T305 | −1.046 | <0.01 | −1.041 | 0.026 | −1.023 | 0.034 |
| CCND1 | P24385 | T286 | - | - | 1.071 | 0.015 | - | - |
| EGFR | P00533 | Y1069 | - | - | 1.032 | 0.041 | - | - |
| EP300 | Q09472 | S89 | - | - | - | - | 1.052 | 0.031 |
| GRB2 | P62993 | Y209 | - | - | −1.063 | 0.02 | 1.044 | 0.017 |
| IL12B | P29460 | Y314 | - | - | −1.048 | 0.03 | - | - |
| IL6ST | P40189 | Y915/Y759 | 1.058 | 0.026 | −1.039 | 0.03 | 1.041 | 0.039 |
| JAK1 | P23458 | Y1034 | - | - | 1.098 | <0.01 | - | - |
| JAK2 | O60674 | Y1007 | −1.059 | 0.027 | −1.042 | <0.01 | - | - |
| MTOR | P42345 | S2481 | 1.061 | 0.042 | −1.058 | 0.01 | −1.041 | 0.036 |
| PDGFRA | P16234 | Y720 | - | - | 1.127 | <0.01 | −1.064 | <0.01 |
| PDGFRB | P09619 | Y579 | −1.115 | <0.01 | −1.061 | 0.019 | −1.061 | 0.046 |
| PIK3CB | P42338 | Y425/S1070 | 1.086 | <0.01 | - | - | −1.049 | 0.022 |
| PIK3R1 | P27986 | S608/Y467 | 1.058 | 0.028 | −1.025 | 0.045 | −1.076 | <0.01 |
| PIK3R2 | O00459 | Y365 | 1.052 | 0.038 | −1.039 | 0.04 | - | - |
| PIM1 | P11309 | S189 | - | - | −1.046 | <0.01 | −1.043 | 0.036 |
| RAF1 | P04049 | S259 | 1.053 | 0.03 | −1.048 | 0.048 | - | - |
| SOCS3 | O14543 | Y204 | −1.069 | <0.01 | −1.052 | <0.01 | - | - |
| SOS1 | Q07889 | S1193 | - | - | - | - | 1.036 | 0.029 |
| STAM2 | O75886 | Y371 | - | - | - | - | −1.057 | 0.017 |
| STAT1 | P42224 | Y701 | 1.057 | 0.048 | 1.060 | 0.033 | 1.034 | 0.034 |
| STAT3 | P40763 | Y705/S727 | 1.067 | 0.015 | 1.078 | 0.031 | 1.046 | 0.044 |
| STAT5B | P51692 | Y699 | −1.071 | <0.01 | 1.061 | <0.01 | 1.087 | <0.01 |
| Chemokine Signaling Pathway | |||
|---|---|---|---|
| Proteins | Day 4 | Day 6 | Day 10 |
| AKT3 | ↓ | ↓ | ↓ |
| ARRB1 | ↓ | ↓ | - |
| ARRB2 | - | ↓ | - |
| CHUK | - | ↑ | - |
| CRK | - | - | ↑ |
| CRKL | ↓ | - | - |
| CRR2 | ↑ | - | - |
| GRB2 | - | ↓ | ↑ |
| GRK5 | ↓ | ↓ | - |
| GSK3A | ↓ | ↑ | ↓ |
| GSK3B | ↓ | ↑ | ↑ |
| JAK2 | ↓ | ↓ | - |
| LYN | - | ↑ | - |
| MAP2K1 | ↑ | - | - |
| NFKB1 | - | ↑ | - |
| NFKBIA | - | ↑ | ↓ |
| PAK1 | ↓ | - | - |
| PIK3R1 | ↑ | ↓ | ↓ |
| PIK3R2 | ↑ | ↓ | - |
| PRKCD | ↓ | ↓ | - |
| PTK2 | ↑ | ↑ | ↓ |
| PTK2B | ↑ | ↑ | ↑ |
| PXN | ↓ | ↑ | ↓ |
| RAF1 | ↑ | ↓ | - |
| SHC1 | - | ↑ | - |
| SHC3 | - | ↑ | - |
| SRC | - | ↓ | ↑ |
| STAT1 | ↑ | ↑ | ↑ |
| STAT3 | ↑ | ↑ | ↑ |
| STAT5B | ↓ | ↑ | ↑ |
| T Cell Receptor Signaling Pathway | |||
|---|---|---|---|
| Proteins | Day 4 | Day 6 | Day 10 |
| AKT3 | ↓ | ↓ | ↓ |
| CHUK | - | ↓ | - |
| FOS | - | ↑ | - |
| FYN | ↓ | ↓ | - |
| GRB2 | - | ↓ | ↑ |
| GSK3B | ↓ | ↑ | ↑ |
| JUN | ↓ | ↓ | - |
| LCK | - | ↑ | - |
| MAP2K1 | ↑ | - | - |
| MAP2K2 | - | ↓ | ↑ |
| MAP3K7 | ↓ | ↑ | ↑ |
| NFATC1 | ↑ | - | - |
| NFATC2 | - | ↓ | - |
| NFATC3 | ↑ | ↓ | ↑ |
| NFKB1 | - | ↑ | - |
| NFKBIA | - | ↑ | ↓ |
| PAK1/2 | ↓ | ↓ | - |
| PDPK1 | ↓ | ↓ | - |
| PIK3R1 | ↑ | ↓ | ↓ |
| PIK3R2 | ↑ | ↓ | ↓ |
| PTPRC | - | ↓ | - |
| RAF1 | ↑ | ↓ | - |
| ZAP70 | - | ↓ | ↓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, A.; Cardoso Dal Pont, G.; Battaglia, M.; Arsenault, R.J.; Kogut, M.H. Role of JAK-STAT Pathway in Broiler Chicks Fed with Chestnut Tannins. Animals 2021, 11, 337. https://doi.org/10.3390/ani11020337
Lee A, Cardoso Dal Pont G, Battaglia M, Arsenault RJ, Kogut MH. Role of JAK-STAT Pathway in Broiler Chicks Fed with Chestnut Tannins. Animals. 2021; 11(2):337. https://doi.org/10.3390/ani11020337
Chicago/Turabian StyleLee, Annah, Gabriela Cardoso Dal Pont, Michele Battaglia, Ryan J. Arsenault, and Michael H. Kogut. 2021. "Role of JAK-STAT Pathway in Broiler Chicks Fed with Chestnut Tannins" Animals 11, no. 2: 337. https://doi.org/10.3390/ani11020337
APA StyleLee, A., Cardoso Dal Pont, G., Battaglia, M., Arsenault, R. J., & Kogut, M. H. (2021). Role of JAK-STAT Pathway in Broiler Chicks Fed with Chestnut Tannins. Animals, 11(2), 337. https://doi.org/10.3390/ani11020337







