Design and Evaluation of a Macroarray for Detection, Identification, and Typing of Viral Hemorrhagic Septicemia Virus (VHSV)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus
2.2. Viral Titration
2.3. Primers and Probes
2.4. The Array System
2.5. RNA Extraction and Quantification
2.6. Reverse Transcription
2.7. TaqMan Real-Time PCR
2.8. One Step Real-Time RT-PCR
2.9. Evaluation of the Efficiency and Reliability of the Array
2.10. Statistical Analysis
3. Results
3.1. Evaluation of the Macroarray Used with RT-qPCR
3.2. Evaluation of the Macroarray Used with qPCR
3.3. Reliability of the Standard Curves for Quantification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mortensen, H.F.; Heuer, O.E.; Lorenzen, N.; Otte, L.; Olesen, N.J. Isolation of viral haemorrhagic septicaemia virus (VHSV) from wild marine fish species in the Baltic Sea, Kattegat, Skagerrak and the North Sea. Virus Res. 1999, 63, 95–106. [Google Scholar] [CrossRef]
- Smail, D.A. Viral haemorrhagic septicaemia. In Fish Diseases and Disorders, Volume 3: Viral, Bacterial and Fungal Infections; Woo, P.T.K., Bruno, D.W., Eds.; CABI Publishing: New York, NY, USA, 1999; pp. 123–147. [Google Scholar]
- Ellis, A.E. Innate host defense mechanism of fish against viruses and bacteria. Dev. Comp. Immunol. 2001, 25, 827–839. [Google Scholar] [CrossRef]
- Einer-Jensen, K.; Ahrens, P.; Forsberg, R.; Lorenzen, N. Evolution of the fish rhabdovirus viral haemorrhagic septicaemia virus. J. Gen. Virol. 2004, 85, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Snow, M.; Bain, N.; Black, J.; Taupin, V.; Cunningham, C.O.; King, J.A.; Skall, H.F.; Raynard, R.S. Genetic population structure of marine viral haemorrhagic septicaemia virus (VHSV). Dis. Aquat. Organ. 2004, 61, 11–12. [Google Scholar] [CrossRef]
- He, M.; Yan, X.C.; Liang, Y.; Sun, X.W.; Teng, C.B. Evolution of the viral hemorrhagic septicemia virus: Divergence, selection and origin. Mol. Phylogenet. Evol. 2014, 77, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.H. Research on the virus of Egtved disease. Ann. N. Y. Acad. Sci. 1965, 126, 422–426. [Google Scholar] [CrossRef]
- Nishizawa, T.; Iida, H.; Takano, R.; Isshiki, T.; Nakajima, K.; Muroga, K. Genetic relatedness among Japanese, American and European isolates of viral hemorrhagic septicemia virus (VHSV) based on partial G and P genes. Dis. Aquat. Organ. 2002, 48, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Guðmundsdóttir, S.; Vendramin, N.; Cuenca, A.; Sigurðardóttir, H.; Kristmundsson, A.; Iburg, T.M.; Olesen, N.J. Outbreak of viral haemorrhagic septicaemia (VHS) in lumpfish (Cyclopterus lumpus) in Iceland caused by VHS virus genotype IV. J. Fish Dis. 2019, 42, 47–62. [Google Scholar] [CrossRef] [Green Version]
- OIE (World Organization for Animal Health). Manual of Diagnostic Tests for Aquatic Animals. 2019. Available online: https://www.oie.int/en/standard-setting/aquatic-manual/access-online/ (accessed on 16 September 2020).
- COUNCIL DIRECTIVE 2006/88/EC (of 24 October 2006). On Animal Health Requirements for Aquaculture Animals and Products Thereof, and on the Prevention and Control of Certain Diseases in Aquatic Animals. Available online: http://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32006L0088&from=EN (accessed on 6 December 2020).
- Kahns, S.; Skall, H.F.; Kaas, R.S.; Korsholm, H.; Bang Jensen, B.; Jonstrup, S.P.; Dodge, M.J.; Einer-Jensen, K.; Stone, D.; Olesen, N.J. European freshwater VHSV genotype Ia isolates divide into two distinct subpopulations. Dis. Aquat. Organ. 2012, 99, 23–35. [Google Scholar] [CrossRef] [Green Version]
- López-Vazquez, C.; Bandín, I.; Dopazo, C.P. RT-Real-time PCR for detection, identification and absolute quantitation of VHSV using different types of standard. Dis. Aquat. Organ. 2015, 114, 99–116. [Google Scholar] [CrossRef] [Green Version]
- Vázquez, D.; López-Vázquez, C.; Skall, H.F.; Mikkelsen, S.S.; Olesen, N.J.; Dopazo, C.P. A novel multiplex RT-qPCR method based on dual-labeled probes suitable for typing all known genotypes of Viral Haemorrhagic Septicaemia Virus. J. Fish Dis. 2015, 39, 467–482. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Walsh, P.S.; Levenson, C.H.; Erlich, H.A. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc. Natl. Acad. Sci. USA 1989, 86, 6230–6234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, S.; Masuta, C.; Sekiguchi, H.; Uehara, T.; Shimura, H.; Maruta, Y. A simple, sensitive, specific detection of mixed infection of multiple plant viruses using macroarray and microtube hybridization. J. Virol. Meth. 2008, 153, 241–244. [Google Scholar] [CrossRef]
- Belàk, S.; Kiss, I.; Viljoen, G.J. New developments in the diagnosis of avian influenza. Rev. Sci. Tech. 2009, 28, 233–243. [Google Scholar] [CrossRef]
- Dopazo, C.P.; Bandín, I. Techniques of diagnosis of fish and shellfish virus and viral diseases. In Safety Analysis of Foods of Animal Origin; Nollet, L., Toldra, F., Eds.; CRC Press: Boca Ratón, FL, USA, 2011; Chapter 18; pp. 531–576. ISBN 978-1-4398-4817-3. [Google Scholar]
- Ries, R.; Beer, M.; Hoffmann, B. BlueTYPE—A low density TaqMan-RT-qPCR array for the identification of all 24 classical Bluetongue virus serotypes. J. Virol. Meth. 2020, 282, 113881. [Google Scholar] [CrossRef]
- Hasan, M.R.; Al Mana, H.; Young, V.; Tang, P.; Thomas, E.; Tan, R.; Tilley, P. A novel real-time PCR assay panel for detection of common respiratory pathogens in a convenient, strip-tube array format. J. Virol. Meth. 2019, 265, 42–48. [Google Scholar] [CrossRef]
- Venter, M.; Zaayman, D.; van Niekerk, S.; Stivaktas, V.; Goolab, S.; Weyer, J.; Paweska, J.T.; Swanepoel, R. Macroarray assay for differential diagnosis of meningoencephalitis in southern Africa. J. Clin. Virol. 2014, 60, 50–56. [Google Scholar] [CrossRef]
- Lievens, B.; Frans, I.; Heusdens, C.; Justé, A.; Jonstrup, S.P.; Lieffrig, F.; Willems, K.A. Rapid detection and identification of viral and bacterial fish pathogens using a DNA array-based multiplex assay. J. Fish. Dis. 2011, 34, 861–875. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, Y.; Jin, R.; Hao, G. A universal random DNA amplification and labeling strategy for microarray to detect multiple pathogens of aquatic animals. J. Virol. Meth. 2020, 275, 113761. [Google Scholar] [CrossRef] [PubMed]
- Lien, K.-Y.; Lee, S.-H.; Tsai, T.-J.; Chen, T.-Y.; Lee, G.-B. A microfluidic-based system using reverse transcription polymerase chain reactions for rapid detection of aquaculture diseases. Microfluid. Nanofluid. 2009, 7, 795–806. [Google Scholar] [CrossRef]
- Le Berre, M.; de Kinkelin, P.; Metzger, A. Identification serologique des Rhabdovirus des Salmonide´s. Bull. Off. Internat. Epizoot. 1977, 87, 391–393. [Google Scholar]
- King, J.A.; Snow, M.; Smail, D.A.; Raynard, R.S. Distribution of viral haemorrhagic septicaemia virus in wild fish species of the North Sea, north east Atlantic Ocean and Irish Sea. Dis. Aquat. Organ. 2001, 47, 81–86. [Google Scholar] [CrossRef]
- Brunson, R.; True, K.; Yancey, J. VHS virus isolated at Makah national fish hatchery. Am. Fish. Soc. Fish. Health Newslett. 1989, 17, 3–4. [Google Scholar]
- Groocock, G.H.; Getchell, R.G.; Wooster, G.A.; Britt, K.L.; Batts, W.N.; Winton, J.R.; Casey, R.N.; Casey, J.W.; Bowser, P.R. Detection of viral hemorrhagic septicemia in round gobies in New York State (USA) waters of Lake Ontario and the St. Lawrence River. Dis. Aquat. Organ. 2007, 76, 187–192. [Google Scholar] [CrossRef] [Green Version]
- López-Vázquez, C.; Bandín, I.; Panzarin, V.; Toffan, A.; Cuenca, A.; Olesen, N.J.; Dopazo, C.P. Steps of the replication cycle of the viral haemorrhagic septicaemia virus (VHSV) affecting its virulence on fish. Animals 2020, 10, 2264. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Dopazo, C.P.; Bandín, I.; López-Vazquez, C.; Lamas, J.; Noya, M.; Barja, J.L. Isolation of viral hemorrhagic septicemia virus from Greenland halibut Reinhardtius hippoglossoides caught in the Flemish Cap. Dis. Aquat. Organ. 2002, 50, 171–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyers, T.R. A Reo-like virus isolated from juvenile American oysters (Crassostrea virginica). J. Gen. Virol. 1979, 43, 203–212. [Google Scholar] [CrossRef]
- Miller, M.B.; Tang, Y.-W. Basic Concepts of Microarrays and Potential Applications in Clinical Microbiology. Clin. Microb. Rev. 2009, 22, 611–633. [Google Scholar] [CrossRef] [Green Version]
- Kabir, M.S. Molecular methods for detection of pathogenic viruses of respiratory tract-A review. Asian Pac. J. Trop. Biomed. 2018, 8, 237–244. [Google Scholar] [CrossRef]
- Tian, M.; Tian, Y.; Li, Y.; Lu, H.; Li, X.; Li, C.; Xue, F.; Jin, N. Microarray Multiplex Assay for the Simultaneous Detection and Discrimination of Influenza A and Influenza B Viruses. Indian J. Microbiol. 2014, 54, 211–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tung, H.Y.; Chen, W.C.; Ou, B.R.; Yeh, J.Y.; Cheng, Y.H.; Tsng, P.H.; Hsu, M.H.; Tsai, M.S.; Liang, Y.C. Simultaneous detection of multiple pathogens by multiplex PCR coupled with DNA biochip hybridization. Lab. Anim. 2018, 52, 186–195. [Google Scholar] [CrossRef]
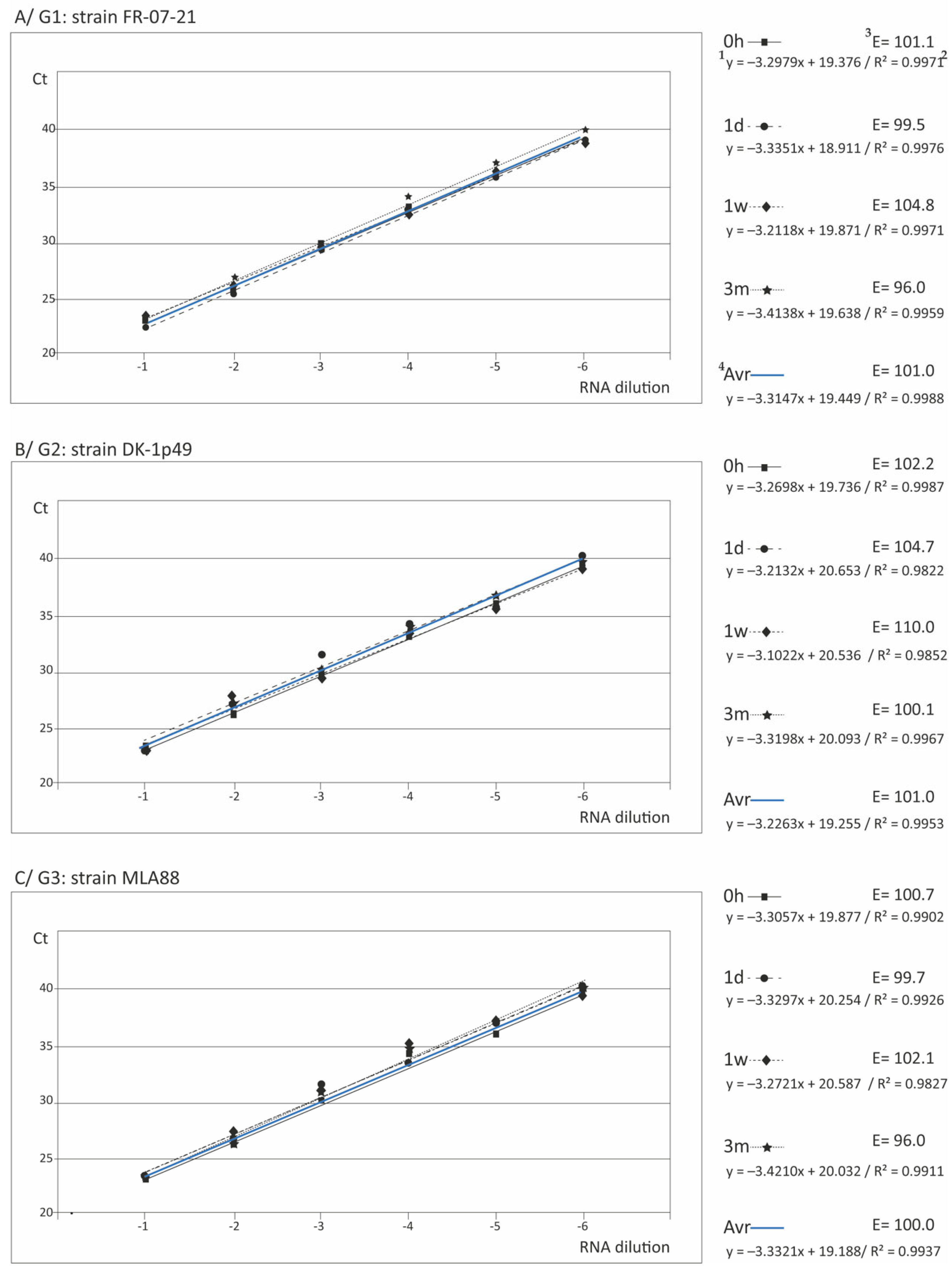
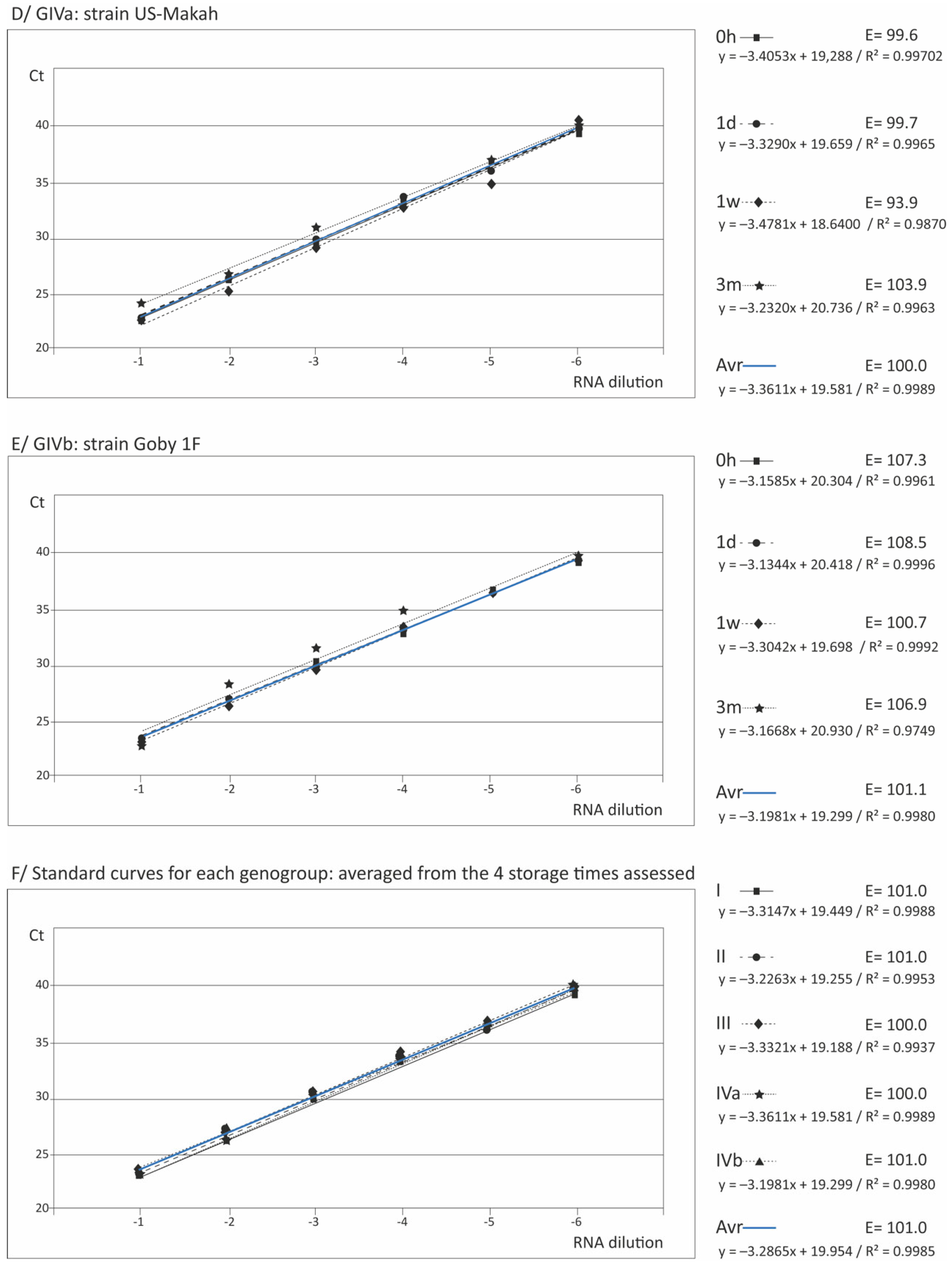


| Strg 1 | a/ RT-qPCR | b/ qPCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 G I. II & III | G. IV | G I. II & III | G. IV | ||||||||
| 0 h | 2 Dil | 10−6 | 10−6 | Dil | 10−7 | 10−6 | |||||
| 3 DL | 5 TCID50/mL | 50 TCID50/mL | LD | 0.5 TCID50/mL | 50 TCID50/mL | ||||||
| 4 Ct | 39.21 ± 0.36 | 39.03 ± 0.23 | Ct | 38.65 ± 0.86 | 36.72 ± 0.47 | ||||||
| 5 E | [101.0-102.2] 101.30 ± 0.79 | [99.6-107.3] 103.45 ± 5.44 | E | [98.6-100.7] 99.70 ± 1.05 | [98.6-105.4] 102.0 ± 4.81 | ||||||
| 6 R2 | 0.9953 ± 0.0045 | 0.9966 ± 0.0006 | R2 | 0.9963 ± 0.0019 | 0.9970 ± 0.0037 | ||||||
| 1 d | Dil | 10−6 | 10−6 | Dil | 10−7 | 10−6 | |||||
| DL | 5 TCID50/mL | 50 TCID50/mL | LD | 0.5 TCID50/mL | 50 TCID50/mL | ||||||
| Ct | 39.61 ± 0.86 | 39.38 ± 0.37 | Ct | 38.48 ± 1.11 | 36.42 ± 1.10 | ||||||
| E | [99.5-104.7] 101.30 ± 2.98 | [99.7-108.5] 104.10 ± 6.22 | E | [99.2-100.8] 97.90 ± 2.81 | [95.1-109.0] 102.05 ± 9.83 | ||||||
| R2 | 0.9908 ± 0.0078 | 0.9980 ± 0.0022 | R2 | 0.9967 ± 0.0022 | 0.9985 ± 0.0008 | ||||||
| 1 w | Dil | 10−6 | 10−6 | Dil | 10−7 | 10−6 | |||||
| DL | 5 TCID50/mL | 50 TCID50/mL | LD | 0.5 TCID50/mL | 50 TCID50/mL | ||||||
| Ct | 39.16 ± 0.16 | 37.46 ± 3.37 | Ct | 38.87 ± 0.84 | 37.23 ± 0.32 | ||||||
| E | [102.1-110.0] 105.33 ± 4.52 | [93.9-100.7] 97.30 ± 4.81 | E | [95.0-99.0] 96.47 ± 2.20 | [97.3-106.1] 101.70 ± 6.22 | ||||||
| R2 | 0.9883 ± 0.0077 | 0.9931 ± 0.0086 | R2 | 0.9979 ± 0.0009 | 0.9997 ± 0.0055 | ||||||
| 3 m | Dil | 10−6 | 10−6 | Dil | 10−7 | 10−6 | |||||
| DL | 5 TCID50/mL | 50 TCID50/mL | LD | 0.5 TCID50/mL | 50 TCID50/mL | ||||||
| Ct | 39.75 ± 0.03 | 40.11 ± 0.03 | Ct | 38.85 ± 0.97 | 37.68 ± 0.97 | ||||||
| E | [96.0-100.1] 97.47 ± 2.29 | [103.9-106.9] 105.40 ± 2.12 | E | [95.2-100.6] 98.20 ± 2.75 | [95.4-102.0] 98.7 ± 4.67 | ||||||
| R2 | 0.9996 ± 0.0030 | 0.9856 ± 0.1513 | R2 | 0.9961 ± 0.0013 | 0.9969 ± 0.0001 | ||||||
| RT-qPCR | qPCR | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GI | GI | |||||||||||||||
| 0 h | 1 d | 1 w | 3 m | 0 h | 1 d | 1 w | 3 m | |||||||||
| 3 GI | 0 h | - | 0.6870 1 0.0572 2 | 0.4279 0.2960 | 0.2330 0.0003 | - | 0.3392 0.4122 | 0.0880 0.4912 | 0.0913 0.6363 | |||||||
| 1 d | - | 0.1999 0.0027 * | 0.3355 <0.0001 * | - | 0.4671 0.9243 | 0.3581 0.2913 | ||||||||||
| 1 w | - | 0.0473 * | - | 0.7441 0.3262 | ||||||||||||
| 3 m | - | - | ||||||||||||||
| GII | GII | |||||||||||||||
| 0 h | 1 d | 1 w | 3 m | 0 h | 1 d | 1 w | 3 m | |||||||||
| GII | 0 h | - | 0.7007 0.0062 * | 0.2154 0.2935 | 0.6276 0.0041 * | - | 0.1941 0.2514 | 0.1730 0.1646 | 0.9426 0.0054 * | |||||||
| 1 d | - | 0.4612 0.1647 | 0.4614 0.4454 | - | 0.8651 0.7263 | 0.1669 0.0917 | ||||||||||
| 1 w | - | 0.1144 0.3423 | - | 0.1499 0.2193 | ||||||||||||
| 3 m | - | - | ||||||||||||||
| GIII | GIII | |||||||||||||||
| 0 h | 1 d | 1 w | 3 m | 0 h | 1 d | 1 w | 3 m | |||||||||
| GIII | 0 h | - | 0.8874 0.1127 | 0.8706 0.0942 | 0.4591 0.0396 * | - | 0.7884 0.5733 | 0.9183 0.1650 | 0.7878 0.9543 | |||||||
| 1 d | - | 0.7924 0.7207 | 0.5924 0.7339 | - | 0.7364 0.1266 | 0.9692 0.6456 | ||||||||||
| 1 w | - | 0.4752 0.9250 | - | 0.7264 0.2148 | ||||||||||||
| 3 m | - | - | ||||||||||||||
| GIVa | GIVa | |||||||||||||||
| 0 h | 1 d | 1 w | 3 m | 0 h | 1 d | 1 w | 3 m | |||||||||
| GIVa | 0 h | - | 0.5133 0.5999 | 0.6104 0.1040 | 0.1462 0.0002 * | - | 0.2099 0.1683 | 0.6969 0.4638 | 0.3377 0.4003 | |||||||
| 1 d | - | 0.3190 0.0569 | 0.4383 0.0013 * | - | 0.4641 0.0467 * | 0.9200 0.0369 * | ||||||||||
| 1 w | - | 0.1061 <0.0001 * | - | 0.5878 0.8919 | ||||||||||||
| 3 m | - | - | ||||||||||||||
| GIVb | GIVb | |||||||||||||||
| 0 h | 1 d | 1 w | 3 m | 0 h | 1 d | 1 w | 3 m | |||||||||
| GIVb | 0 h | - | 0.8433 0.8850 | 0.1548 0.5818 | 0.9618 0.0262 * | - | 0.4392 0.0992 | 0.9016 0.2244 | 0.4585 <0.0001 * | |||||||
| 1 d | - | 0.1433 0.5257 | 0.8549 0.0418 * | - | 0.6623 0.0210* | 0.1596 <0.0001 * | ||||||||||
| 1 w | - | 0.4018 0.0100 * | - | 0.5123 <0.0001 * | ||||||||||||
| 3 m | - | - | ||||||||||||||
| RT-qPCR | qPCR | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 Genogr. | GI | GII | GIII | GIVa | GIVb | GI | GII | GIII | GIVa | GIVb | ||||||||||
| GI | - | 0.2598 1 | 0.7936 | 0.4375 | 0.0689 | - | 0.9678 | 0.2650 | 0.8943 | 0.0109 | ||||||||||
| - | 0.0006 2,* | <0.0001 * | 0.0174 * | 0.0003 * | - | 0.0072 * | <0.0001 | <0.0001 | ** | |||||||||||
| GII | - | - | 0.2880 | 0.1132 | 0.7380 | - | - | 0.1951 | 0.9039 | 0.0036 | ||||||||||
| - | - | 0.0960 | 0.1593 | 0.6605 | - | - | <0.0001 | <0.0001 | ** | |||||||||||
| GIII | - | - | - | 0.7584 | 0.1409 | - | - | - | 0.2150 | 0.1161 | ||||||||||
| - | - | - | 0.0029 * | 0.0300 * | - | - | - | <0.0001 | <0.0001 | |||||||||||
| GIVa | - | - | - | - | 0.0280 * | - | - | - | - | 0.0059 | ||||||||||
| - | - | - | - | ** | - | - | - | - | ** | |||||||||||
| GIVb | - | - | - | - | - | - | - | - | - | - | ||||||||||
| - | - | - | - | - | - | - | - | - | - | |||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Vázquez, C.; Bandín, I.; Dopazo, C.P. Design and Evaluation of a Macroarray for Detection, Identification, and Typing of Viral Hemorrhagic Septicemia Virus (VHSV). Animals 2021, 11, 841. https://doi.org/10.3390/ani11030841
López-Vázquez C, Bandín I, Dopazo CP. Design and Evaluation of a Macroarray for Detection, Identification, and Typing of Viral Hemorrhagic Septicemia Virus (VHSV). Animals. 2021; 11(3):841. https://doi.org/10.3390/ani11030841
Chicago/Turabian StyleLópez-Vázquez, Carmen, Isabel Bandín, and Carlos P. Dopazo. 2021. "Design and Evaluation of a Macroarray for Detection, Identification, and Typing of Viral Hemorrhagic Septicemia Virus (VHSV)" Animals 11, no. 3: 841. https://doi.org/10.3390/ani11030841
APA StyleLópez-Vázquez, C., Bandín, I., & Dopazo, C. P. (2021). Design and Evaluation of a Macroarray for Detection, Identification, and Typing of Viral Hemorrhagic Septicemia Virus (VHSV). Animals, 11(3), 841. https://doi.org/10.3390/ani11030841







