Processing Matters in Nutrient-Matched Laboratory Diets for Mice—Microbiome
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Post Mortem Sampling
2.3. Metagenomic DNA Extraction
2.4. Data Processing
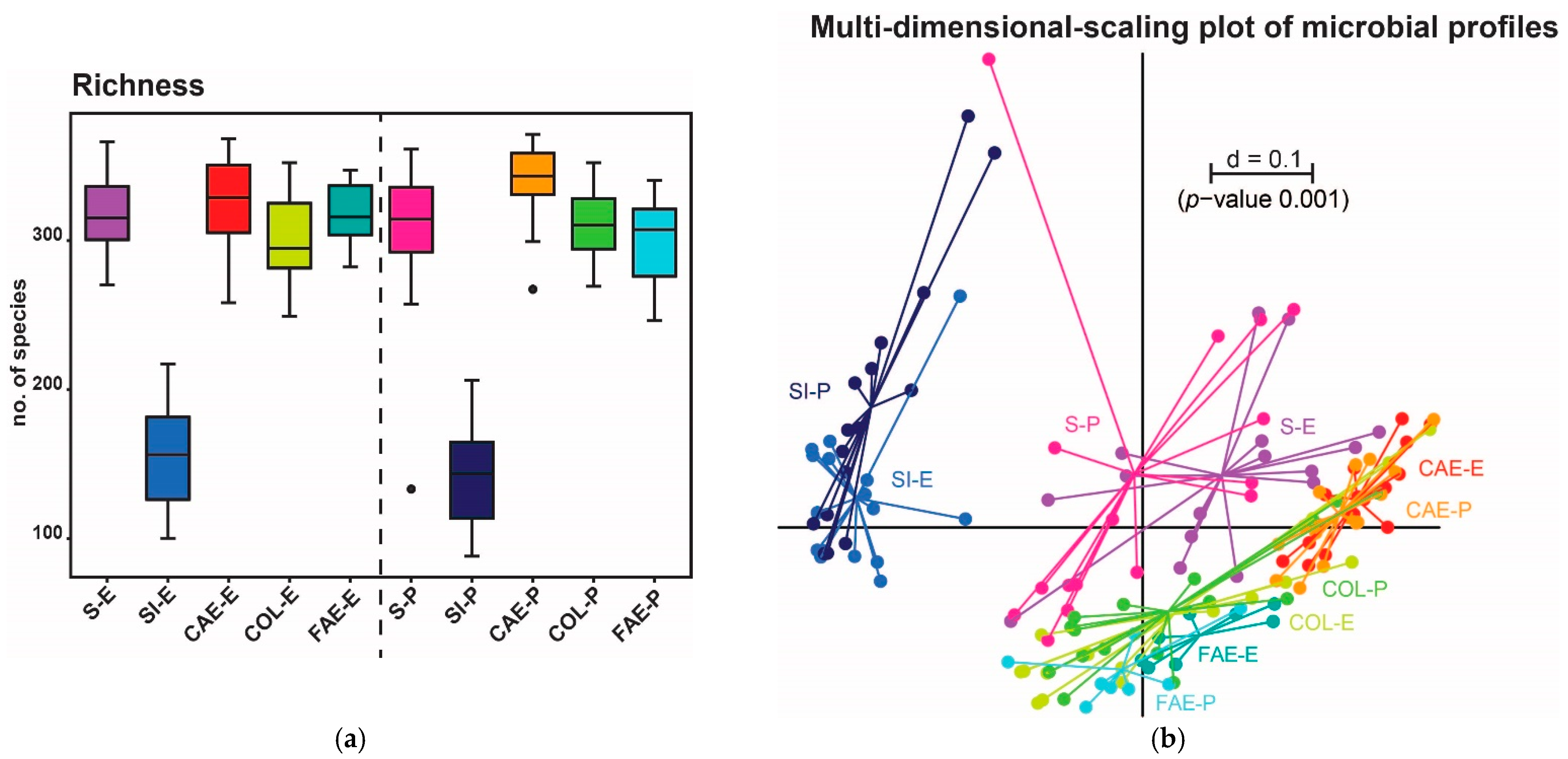
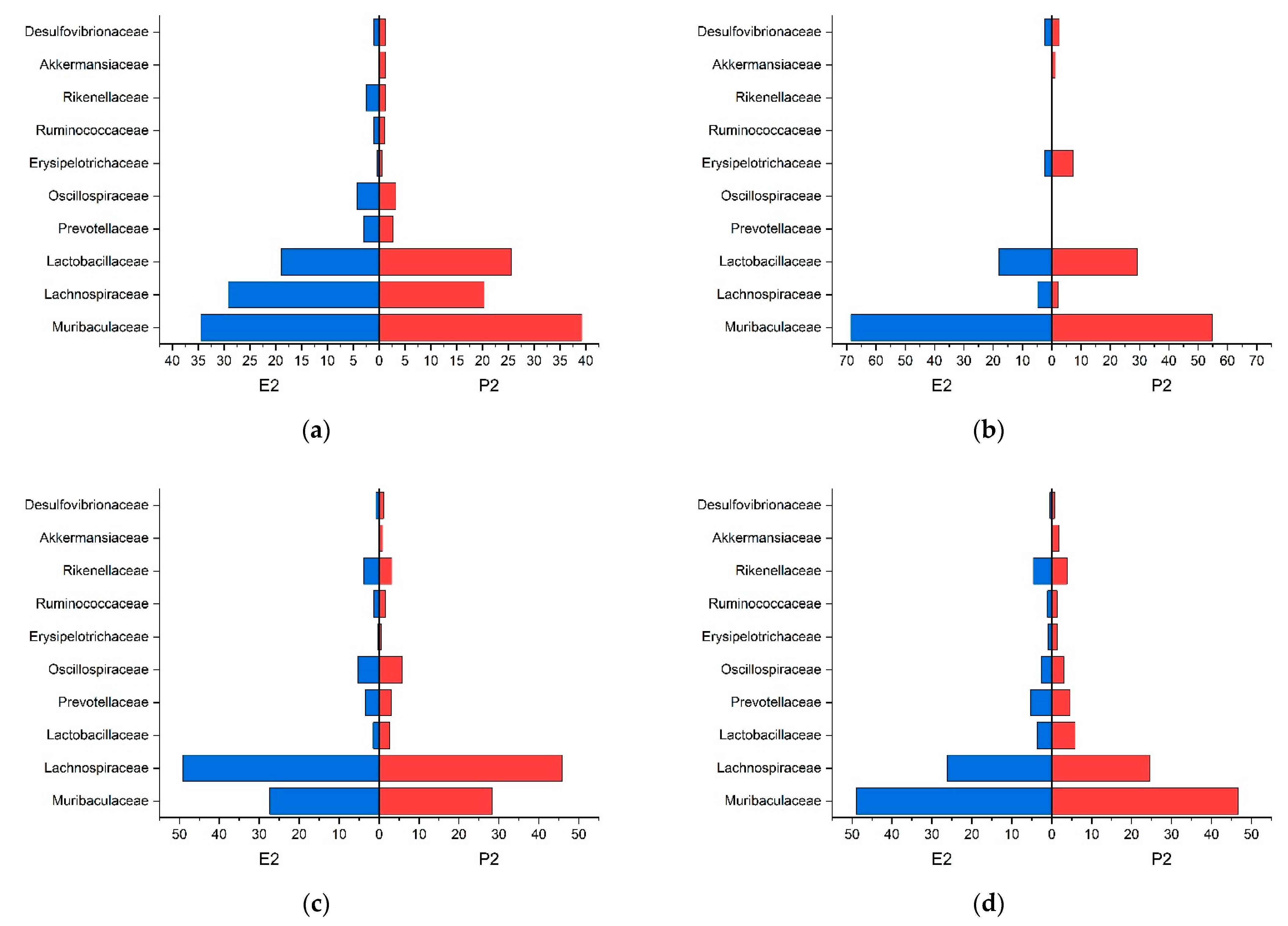
3. Results
3.1. Trial 1
3.2. Trial 2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; A Relman, D. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Clavel, T.; Desmarchelier, C.; Haller, D.; Gérard, P.; Rohn, S.; Lepage, P.; Daniel, H. Intestinal microbiota in metabolic diseases. Gut Microbes 2014, 5, 544–551. [Google Scholar] [CrossRef]
- Cummings, J.H.; Beatty, E.R.; Kingman, S.M.; Bingham, S.A.; Englyst, H.N. Digestion and physiological properties of resistant starch in the human large bowel. Br. J. Nutr. 1996, 75, 733–747. [Google Scholar] [CrossRef]
- Bouskra, D.; Brézillon, C.; Bérard, M.; Werts, C.; Varona, R.; Boneca, I.G.; Eberl, G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nat. Cell Biol. 2008, 456, 507–510. [Google Scholar] [CrossRef]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef]
- Banasaz, M.; Norin, E.; Holma, R.; Midtvedt, T. Increased Enterocyte Production in Gnotobiotic Rats Mono-Associated with Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2002, 68, 3031–3034. [Google Scholar] [CrossRef]
- Hooper, L.V.; Stappenbeck, T.S.; Hong, C.V.; Gordon, J.I. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 2003, 4, 269–273. [Google Scholar] [CrossRef]
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic Bacteria Direct Expression of an Intestinal Bactericidal Lectin. Science 2006, 313, 1126–1130. [Google Scholar] [CrossRef]
- Salzman, N.H.; Underwood, M.A.; Bevins, C.L. Paneth cells, defensins, and the commensal microbiota: A hypothesis on intimate interplay at the intestinal mucosa. Semin. Immunol. 2007, 19, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Gordon, H.A.; Wostmann, B.S.; Bruckner-Kardoss, E. Effects of Microbial Flora on Cardiac Output and Other Elements of Blood Circulation. Exp. Biol. Med. 1963, 114, 301–304. [Google Scholar] [CrossRef]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.-N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef]
- Sudo, N. Stress and gut microbiota: Does postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response? Int. Congr. Ser. 2006, 1287, 350–354. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Sartor, R.B. Microbial Influences in Inflammatory Bowel Diseases. Gastroenterology 2008, 134, 577–594. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, H.; Liu, Z. Interplay of intestinal microbiota and mucosal immunity in inflammatory bowel disease: A relationship of frenemies. Ther. Adv. Gastroenterol. 2020, 13. [Google Scholar] [CrossRef]
- E Moore, W.; Moore, L.H. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 1995, 61, 3202–3207. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Shanahan, F.; Clune, Y.; Collins, J.K.; O’Sullivan, G.C.; O’Riordan, M.; Holmes, E.; Wang, Y.; Marchesi, J.R. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ. Microbiol. 2008, 10, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nat. Cell Biol. 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Hansen, A.K.; Ling, F.; Kaas, A.; Funda, D.P.; Farlov, H.; Buschard, K. Diabetes preventive gluten-free diet decreases the number of caecal bacteria in non-obese diabetic mice. Diabetes/Metab. Res. Rev. 2006, 22, 220–225. [Google Scholar] [CrossRef]
- Wen, L.; Ley, R.E.; Volchkov, P.Y.; Stranges, P.B.; Avanesyan, L.; Stonebraker, A.C.; Hu, C.; Wong, F.S.; Szot, G.L.; Bluestone, J.A.; et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nat. Cell Biol. 2008, 455, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Rausch, P.; Basic, M.; Batra, A.; Bischoff, S.C.; Blaut, M.; Clavel, T.; Gläsner, J.; Gopalakrishnan, S.; Grassl, G.A.; Günther, C.; et al. Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int. J. Med Microbiol. 2016, 306, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Sajilata, M.G.; Singhal, R.S.; Kulkarni, P.R. Resistant Starch? A Review. Compr. Rev. Food Sci. Food Saf. 2006, 5, 1–17. [Google Scholar] [CrossRef]
- Copeland, L.; Blazek, J.; Salman, H.; Tang, M.C. Form and functionality of starch. Food Hydrocoll. 2009, 23, 1527–1534. [Google Scholar] [CrossRef]
- Hoover, R. The Impact of Heat-Moisture Treatment on Molecular Structures and Properties of Starches Isolated from Different Botanical Sources. Crit. Rev. Food Sci. Nutr. 2010, 50, 835–847. [Google Scholar] [CrossRef]
- Englyst, H.N.; Kingman, S.M.; Cummings, J.H. Classification and measurement of nutritionally important starch fractions. Eur. J. Clin. Nutr. 1992, 46, S33–S50. [Google Scholar]
- Englyst, H.N.; Kingman, S.M.; Hudson, G.J.; Cummings, J.H. Measurement of resistant starch in vitro and in vivo. Br. J. Nutr. 1996, 75, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Berry, C. Resistant starch: Formation and measurement of starch that survives exhaustive digestion with amylolytic enzymes during the determination of dietary fibre. J. Cereal Sci. 1986, 4, 301–314. [Google Scholar] [CrossRef]
- Wang, S.; Copeland, L. Molecular disassembly of starch granules during gelatinization and its effect on starch digestibility: A review. Food Funct. 2013, 4, 1564–1580. [Google Scholar] [CrossRef]
- Kienzle, E. Carbohydrate metabolism of the cat 2. Digestion of starch. J. Anim. Physiol. Anim. Nutr. 1993, 69, 102–114. [Google Scholar] [CrossRef]
- Kienzle, E.; Pohlenz, J.; Radicke, S. Morphology of Starch Digestion in The Horse. J. Veter. Med. Ser. A 1997, 44, 207–221. [Google Scholar] [CrossRef]
- Böswald, L.; Wenderlein, J.; Straubinger, R.; Ulrich, S.; Kienzle, E. Processing Matters in Nutrient-Matched Laboratory Diets for Mice—Energy and Nutrient Digestibility. Animals 2021, 11, 523. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-J.; Lim, H.S.; Lim, S.-T. Effect of partial gelatinization and retrogradation on the enzymatic digestion of waxy rice starch. J. Cereal Sci. 2006, 43, 353–359. [Google Scholar] [CrossRef]
- Kleessen, B.; Stoof, G.; Proll, J.; Schmiedl, D.; Noack, J.; Blaut, M. Feeding resistant starch affects fecal and cecal microflora and short-chain fatty acids in rats. J. Anim. Sci. 1997, 75, 2453–2462. [Google Scholar] [CrossRef]
- Tachon, S.; Zhou, J.; Keenan, M.; Martin, R.; Marco, M.L. The intestinal microbiota in aged mice is modulated by dietary resistant starch and correlated with improvements in host responses. FEMS Microbiol. Ecol. 2012, 83, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, E.; Young, W.; Reichert-Grimm, V.; Weis, S.; Riedel, C.U.; Rosendale, D.; Stoklosinski, H.; Hunt, M.; Egert, M. In Vivo Assessment of Resistant Starch Degradation by the Caecal Microbiota of Mice Using RNA-Based Stable Isotope Probing—A Proof-of-Principle Study. Nutrients 2018, 10, 179. [Google Scholar] [CrossRef] [PubMed]
- Desmarchelier, C.; Ludwig, T.; Scheundel, R.; Rink, N.; Bader, B.L.; Klingenspor, M.; Daniel, H. Diet-induced obesity in ad libitum-fed mice: Food texture overrides the effect of macronutrient composition. Br. J. Nutr. 2012, 109, 1518–1527. [Google Scholar] [CrossRef]
- Reitmeier, S.; Kiessling, S.; Neuhaus, K.; Haller, D. Comparing Circadian Rhythmicity in the Human Gut Microbiome. STAR Protoc. 2020, 1, 100148. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Kläring, K.; Heinzmann, S.S.; Platz, S.; Scholz, B.; Engel, K.-H.; Schmitt-Kopplin, P.; Haller, D.; Rohn, S.; Skurk, T.; et al. Gut metabolites and bacterial community networks during a pilot intervention study with flaxseeds in healthy adult men. Mol. Nutr. Food Res. 2015, 59, 1614–1628. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Joseph, D.; Kapfhammer, M.; Giritli, S.; Horn, M.; Haller, D.; Clavel, T. IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci. Rep. 2016, 6, srep33721. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved Error-Correction for Illumina 16S and Its Amplicon Sequencing. bioRxiv 2016, 081257. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Fischer, S.; Kumar, N.; Clavel, T. Rhea: A transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 2017, 5, e2836. [Google Scholar] [CrossRef] [PubMed]
- Jost, L. Partitioning diversity into independent alpha and beta components. Ecology 2007, 88, 2427–2439. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B-Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Risely, A. Applying the core microbiome to understand host–microbe systems. J. Anim. Ecol. 2020, 89, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Do, M.H.; Lee, H.-B.; Lee, E.; Park, H.-Y. The Effects of Gelatinized Wheat Starch and High Salt Diet on Gut Microbiota and Metabolic Disorder. Nutrients 2020, 12, 301. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Pukall, R.; Abt, B.; Foesel, B.U.; Meier-Kolthoff, J.P.; Kumar, N.; Bresciani, A.; Martínez, I.; Just, S.; Ziegler, C.; et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat. Microbiol. 2016, 1, 16131. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.A.; Gálvez, E.J.C.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Miyake, S.; Ding, Y.; Soh, M.; Low, A.; Seedorf, H. Cultivation and description of Duncaniella dubosii sp. nov., Duncaniella freteri sp. nov. and emended description of the species Duncaniella muris. Int. J. Syst. Evol. Microbiol. 2020, 70, 3105–3110. [Google Scholar] [CrossRef]
- Ormerod, K.L.; Wood, D.L.A.; Lachner, N.; Gellatly, S.L.; Daly, J.N.; Parsons, J.D.; Dal’Molin, C.G.O.; Palfreyman, R.W.; Nielsen, L.K.; Cooper, M.A.; et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 2016, 4, 36. [Google Scholar] [CrossRef]
- Macfarlane, G.; Gibson, G.R. Formation of glycoprotein degrading enzymes by Bacteroides fragilis. FEMS Microbiol. Lett. 1991, 77, 289–293. [Google Scholar] [CrossRef]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353.e21. [Google Scholar] [CrossRef] [PubMed]
- Do, M.H.; Lee, E.; Oh, M.-J.; Kim, Y.; Park, H.-Y. High-Glucose or -Fructose Diet Cause Changes of the Gut Microbiota and Metabolic Disorders in Mice without Body Weight Change. Nutrients 2018, 10, 761. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Chin, Y.; Chen, X.; Mi, Y.; Xue, C.; Wang, Y.; Tang, Q. The role of gut microbiota in the resistance to obesity in mice fed a high fat diet. Int. J. Food Sci. Nutr. 2019, 71, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Toh, H.; Fukuda, S.; Horikawa, H.; Oshima, K.; Suzuki, T.; Murakami, M.; Hisamatsu, S.; Kato, Y.; Takizawa, T.; et al. Comparative Genome Analysis of Lactobacillus reuteri and Lactobacillus fermentum Reveal a Genomic Island for Reuterin and Cobalamin Production. DNA Res. 2008, 15, 151–161. [Google Scholar] [CrossRef] [PubMed]




| Sample | E1 | P1 | p-Value | |
|---|---|---|---|---|
| Stomach content | Read count | 23,679 | 19,682 | |
| Richness | 313.6 | 264.4 | <0.01 | |
| Small intestinal content | Read count | 21,910 | 19,252 | |
| Richness | 135.3 | 134.4 | 0.92 | |
| Cecal content | Read count | 22,582 | 19,614 | |
| Richness | 333.9 | 278.6 | <0.001 | |
| Colon content | Read count | 21,778 | 20,216 | |
| Richness | 288.6 | 270.4 | 0.98 | |
| Feces | Read count | 20,236 | 20,883 | |
| Richness | 315.3 | 284.0 | 0.43 |
| Sample | E2 | P2 | p-Value | |
|---|---|---|---|---|
| Stomach content | Read count | 23,127 | 23,741 | |
| Richness | 320.4 | 308.1 | 0.84 | |
| Small intestinal content | Read count | 22,168 | 21,752 | |
| Richness | 158.8 | 146.2 | 0.36 | |
| Cecal content | Read count | 24,453 | 24,077 | |
| Richness | 330.6 | 342.3 | 0.41 | |
| Colon content | Read count | 23,262 | 22,953 | |
| Richness | 306.0 | 315.9 | 0.45 | |
| Feces | Read count | 21,508 | 24,614 | |
| Richness | 322.3 | 302.5 | 0.37 |
| Sample | E1 (%) | P1 (%) |
|---|---|---|
| Stomach content | 79.3 | 83.5 |
| Small intestinal content | 78.1 | 91.9 |
| Cecal content | 76.5 | 80.9 |
| Colon content | 73.8 | 83.5 |
| Feces | 79.3 | 81.1 |
| Sample | E2 (%) | P2 (%) |
|---|---|---|
| Stomach content | 82.6 | 85.2 |
| Small intestinal content | 91.5 | 86.5 |
| Cecal content | 78.0 | 76.8 |
| Colon content | 78.7 | 77.1 |
| Feces | 77.8 | 75.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wenderlein, J.; Böswald, L.F.; Ulrich, S.; Kienzle, E.; Neuhaus, K.; Lagkouvardos, I.; Zenner, C.; Straubinger, R.K. Processing Matters in Nutrient-Matched Laboratory Diets for Mice—Microbiome. Animals 2021, 11, 862. https://doi.org/10.3390/ani11030862
Wenderlein J, Böswald LF, Ulrich S, Kienzle E, Neuhaus K, Lagkouvardos I, Zenner C, Straubinger RK. Processing Matters in Nutrient-Matched Laboratory Diets for Mice—Microbiome. Animals. 2021; 11(3):862. https://doi.org/10.3390/ani11030862
Chicago/Turabian StyleWenderlein, Jasmin, Linda F. Böswald, Sebastian Ulrich, Ellen Kienzle, Klaus Neuhaus, Ilias Lagkouvardos, Christian Zenner, and Reinhard K. Straubinger. 2021. "Processing Matters in Nutrient-Matched Laboratory Diets for Mice—Microbiome" Animals 11, no. 3: 862. https://doi.org/10.3390/ani11030862
APA StyleWenderlein, J., Böswald, L. F., Ulrich, S., Kienzle, E., Neuhaus, K., Lagkouvardos, I., Zenner, C., & Straubinger, R. K. (2021). Processing Matters in Nutrient-Matched Laboratory Diets for Mice—Microbiome. Animals, 11(3), 862. https://doi.org/10.3390/ani11030862







