The Effect of Infertility on the Liver Structure, Endocrinology, and Gene Network in Japanese Flounder
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Fish Sampling
2.3. Histological Analysis and Hormonal Assays
2.4. RNA Extraction, Library Construction, and Sequencing
2.5. Analysis of Protein-Coding and lncRNA Transcripts
2.6. Analysis of circRNA Transcripts
2.7. miRNA Analysis
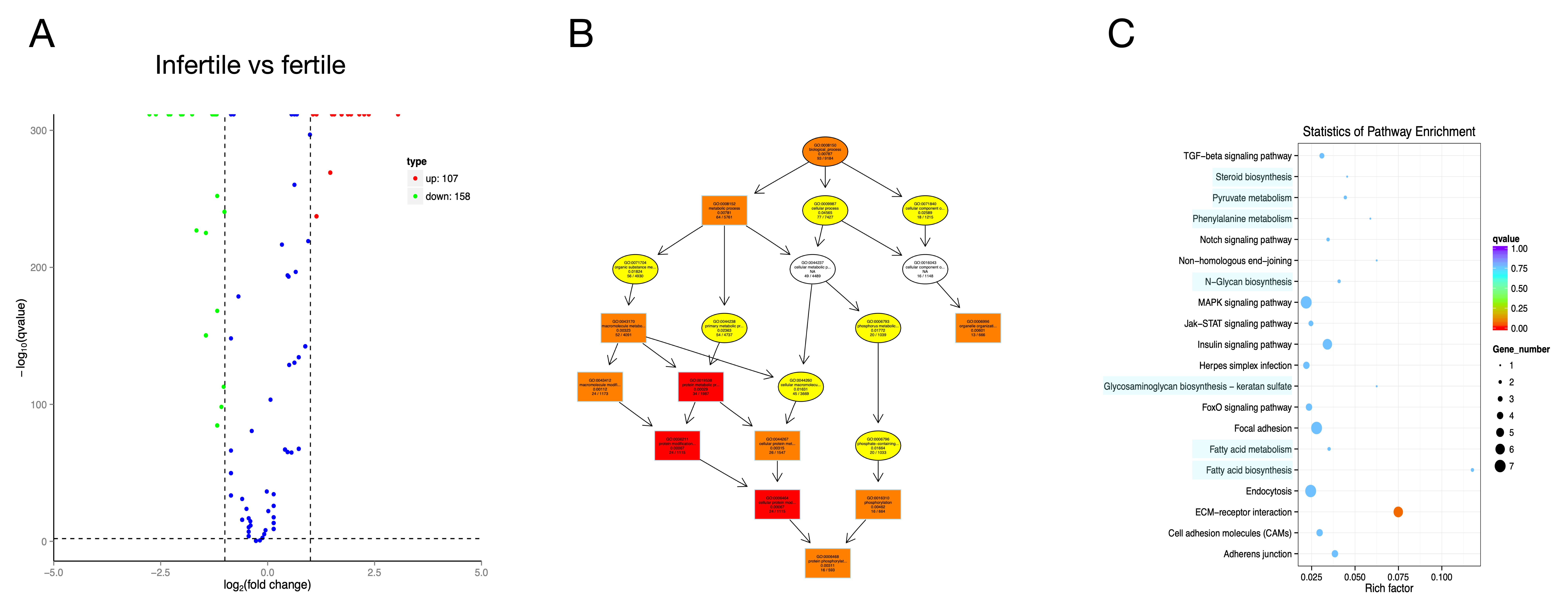
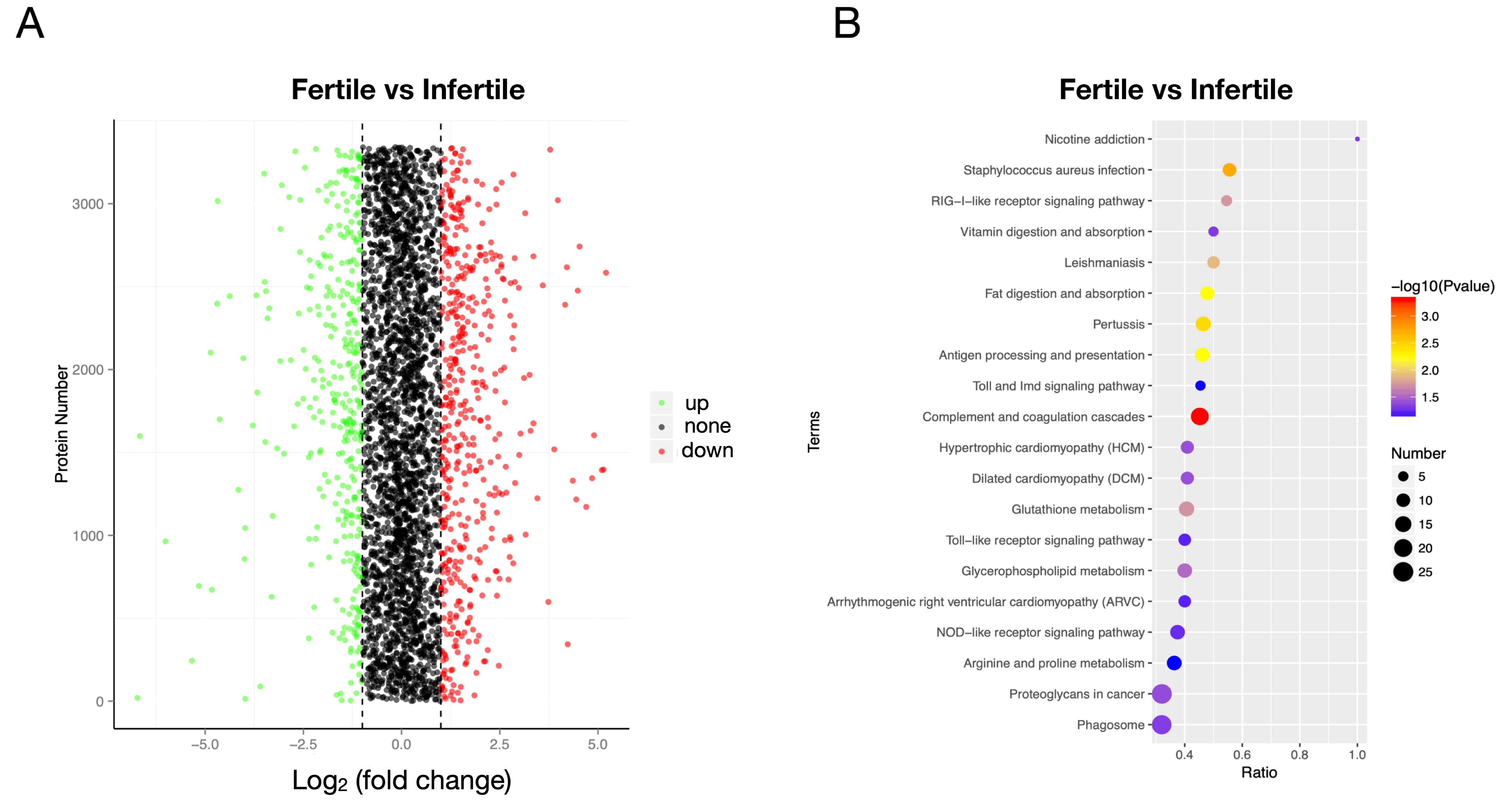
2.8. Functional Enrichment Analysis
2.9. Integrated Analysis for Whole Transcriptomics
2.10. Validation of Differentially Expressed Genes (DEGs) by qRT-PCR
2.11. Proteomic Analysis
2.12. Integrated Transcriptomic and Proteomic Analysis
2.13. Construction of miRNA/lncRNA/circRNA and cor-DEGs-DEPs Genes Relationship Network
3. Results
3.1. Gonad and Liver Histology
3.2. Hormonal Concentrations
3.3. Whole Transcriptomic Analysis
3.4. Construction of ceRNA Network
3.5. Validation of DEGs by qRT-PCR
3.6. Proteome Profile and Expression Differences
3.7. Integrated Analysis of Transcriptomics and Proteomics
3.8. Construction of miRNA/lncRNA/circRNA and cor-DEGs-DEPs Genes Relationship Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Drew, E.M. Infertility in the modern world: Present and future prospects. Am. J. Hum. Biol. 2002, 14, 284–285. [Google Scholar] [CrossRef]
- Ehrmann, D.A.; Barnes, R.B.; Rosenfield, R.L.; Cavaghan, M.K.; Imperial, J. Prevalence of impaired glucose tolerance and diabetes in women with polycystic ovary syndrome. Diabetes Care 1999, 22, 141–146. [Google Scholar] [CrossRef]
- Laven, J.S.; Imani, B.; Eijkemans, M.J.; Fauser, B.C. New approach to polycystic ovary syndrome and other forms of anovulatory infertility. Obstet. Gynecol. Surv. 2002, 57, 755–767. [Google Scholar] [CrossRef]
- van der Helm, D.; Barnhoorn, M.C.; de Jonge-Muller, E.S.; Molendijk, I.; Hawinkels, L.J.; Coenraad, M.J.; van Hoek, B.; Verspaget, H.W. Local but not systemic administration of mesenchymal stromal cells ameliorates fibrogenesis in regenerating livers. J. Cell. Mol. Med. 2019, 23, 6238–6250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Hu, H.; Wan, Y.; Zhang, Y.; Zheng, L.; Hong, Z. Pien Tze Huang Gan Bao ameliorates carbon tetrachloride-induced hepatocyte apoptosis in rats, associated with suppression of p53 activation and oxidative stress. Mol. Med. Rep. 2017, 16, 2611–2619. [Google Scholar] [CrossRef] [Green Version]
- Ferré, L.E.; Medesani, D.A.; García, C.F.; Grodzielski, M.; Rodríguez, E.M. Vitellogenin levels in hemolymph, ovary and hepatopancreas of the freshwater crayfish Cherax quadricarinatus (Decapoda: Parastacidae) during the reproductive cycle. Rev. Biol. Trop. 2012, 60, 253–261. [Google Scholar] [CrossRef] [Green Version]
- Baumann, L.; Holbech, H.; Keiter, S.; Kinnberg, K.L.; Knörr, S.; Nagel, T.; Braunbeck, T. The maturity index as a tool to facilitate the interpretation of changes in vitellogenin production and sex ratio in the Fish Sexual Development Test. Aquat. Toxicol. 2013, 128, 34–42. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, G.; Sun, Z.; Wang, Y.; Hou, J. Mass Production of Doubled Haploids in Japanese Flounder, Paralichthys olivaceus. J. World Aquacult. Soc. 2018, 49, 420–428. [Google Scholar] [CrossRef]
- Bongers, A.; Zandieh-Doulabi, B.; Richter, C.J.; Komen, J. Viable androgenetic YY genotypes of common carp (Cyprinus carpio L.). J. Hered. 1999, 90, 195–198. [Google Scholar] [CrossRef] [Green Version]
- Müller-Belecke, A.; Hörstgen-Schwark, G. Sex determination in tilapia (Oreochromis niloticus) sex ratios in homozygous gynogenetic progeny and their offspring. Aquaculture 1995, 137, 57–65. [Google Scholar] [CrossRef]
- Zhang, X.; Hou, J.; Wang, G.; Jiang, H.; Wang, Y.; Sun, Z.; Jiang, X.; Yu, Q.; Liu, H. Gonadal Transcriptome Analysis in Sterile Double Haploid Japanese Flounder. PLoS ONE 2016, 11, e147668. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, G.; Sun, Z.; Hou, J.; Wang, Y. microRNA-mRNA analysis in pituitary and hypothalamus of sterile Japanese flounder. Mol. Reprod. Dev. 2019, 86, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhong, Z.; Lv, M.; Shu, J.; Tian, Q.; Chen, J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget 2016, 7, 47186–47200. [Google Scholar] [CrossRef]
- Li, N.; Liu, X.L.; Zhang, F.L.; Tian, Y.; Zhu, M.; Meng, L.Y.; Dyce, P.W.; Shen, W.; Li, L. Whole-transcriptome analysis of the toxic effects of zearalenone exposure on ceRNA networks in porcine granulosa cells. Environ. Pollut. 2020, 261, 114007. [Google Scholar] [CrossRef]
- Mahadevan, C.; Krishnan, A.; Saraswathy, G.G.; Surendran, A.; Jaleel, A.; Sakuntala, M. Transcriptome- Assisted Label-Free Quantitative Proteomics Analysis Reveals Novel Insights into Piper nigrum—Phytophthora capsici Phytopathosystem. Front. Plant. Sci. 2016, 7, 785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogl, S.; van Bebber, F.; Dislich, B.; Kuhn, P.H.; Haass, C.; Schmid, B.; Lichtenthaler, S.F. Label-free quantitative analysis of the membrane proteome of Bace1 protease knock-out zebrafish brains. Proteomics 2013, 13, 1519–1527. [Google Scholar] [CrossRef]
- Yamamoto, E. Studies on sex-manipulation and production of cloned populations in hirame, Paralichthys olivaceus (Temminck et Schlegel). Aquaculture 1999, 173, 235–246. [Google Scholar] [CrossRef]
- Shao, C.; Bao, B.; Xie, Z.; Chen, X.; Li, B.; Jia, X.; Yao, Q.; Orti, G.; Li, W.; Li, X.; et al. The genome and transcriptome of Japanese flounder provide insights into flatfish asymmetry. Nat. Genet. 2017, 49, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018, 19, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; et al. Integrated Profiling of MicroRNAs and mRNAs: MicroRNAs Located on Xq27.3 Associate with Clear Cell Renal Cell Carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Enright, A.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. goseq: Gene Ontology testing for RNA-seq datasets. Gene 2010, 8, 1–21. [Google Scholar]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jiang, H.B.; Bao, J.; Zhang, X.Y.; Wang, G.X.; Liu, H.J.; Jiang, L.; Han, Y. Proteomic Analysis of Proteins Related to Homozygotic Sterility Derived from Gynogenesis in the Japanese Flounder, Paralichthys olivaceus. J. World Aquacult. Soc. 2017, 6, 938–946. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; Von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husain, M.; Rasool, K.G.; Tufail, M.; Aldawood, A.S. Molecular characterization, expression pattern and RNAi-mediated silencing of vitellogenin receptor gene in almond moth, Cadra cautella. Insect. Mol. Biol. 2020, 29, 417–430. [Google Scholar] [CrossRef]
- Hiramatsu, N.; Cheek, A.O.; Sullivan, C.V.; Matsubara, T.; Hara, A. Vitellogenesis and endocrine disruption. In Biochemistry and Molecular Biology of Fishes; Mommsen, T.P., Moon, T.W., Eds.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 431–471. [Google Scholar]
- Passantino, L.; Zupa, R.; Pousis, C.; Mylonas, C.C.; Hala, E.; Jirillo, E.; Corriero, A. Increased melanomacrophage centres in the liver of reproductively dysfunctional female greater amberjack Seriola dumerili (Risso, 1810). J. Fish. Dis. 2020, 43, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Wakil, S.J. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry 2002, 28, 4523–4530. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Shi, T.; Zhang, L.; Zhu, P.; Deng, M.; Huang, C.; Hu, T.; Jiang, L.; Li, J. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 2016, 370, 153–164. [Google Scholar] [CrossRef]
- Van Schaftingen, E.; Gerin, I. The glucose-6-phosphatase system. Biochem. J. 2002, 362, 513–532. [Google Scholar] [CrossRef]
- Mueckler, M.; Thorens, B. The SLC2 (GLUT) family of membrane transporters. Mol. Asp. Med. 2013, 34, 121–138. [Google Scholar] [CrossRef] [Green Version]
- Oğuz, A.R.; Ünal, G. In vitro effect of pituitary, interrenal and gonadal hormones on vitellogenin synthesis in primary hepatocyte cultures of Chalcalburnus tarichi. Aquac. Res. 2015, 46, 482–491. [Google Scholar] [CrossRef]
- Takemura, A.; Kim, B.H. Effects of estradiol-17β treatment on in vitro and in vivo synthesis of two distinct vitellogenins in tilapia. Comp. Biochem. Phys. A 2001, 129, 641–651. [Google Scholar] [CrossRef]
- Tyler, C.R.; Sumpter, J.P.; Kawauchi, H.; Swanson, P. Involvement of gonadotropin in the uptake of vitellogenin into vitellogenic oocytes of the rainbow trout, Oncorhynchus mykiss. Gen. Comp. Endocr. 1991, 84, 291–299. [Google Scholar] [CrossRef]
- Liu, Q.; Wen, J.; Ou, Y.; Li, J.E.; Zhou, H.; Tang, Q. Effects of acute handling stress on liver tissue and oxidative stress of juvenile Eleutheronema tetradactylum. South Chnia Fish. Sci. Sci. 2017, 5, 103–109. [Google Scholar]
- Nicoli, S.; Knyphausen, C.P.; Zhu, L.J.; Lakshmanan, A.; Lawson, N.D. miR-221 is required for endothelial tip cell behaviors during vascular development. Dev. Cell 2012, 22, 418–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.D.; Wang, D.D.; Wang, Z.; Wang, Y.B.; Li, G.X.; Sun, G.R.; Tian, Y.D.; Han, R.L.; Li, Z.J.; Jiang, R.R.; et al. Estrogen Abolishes the Repression Role of gga-miR-221-5p Targeting ELOVL6 and SQLE to Promote Lipid Synthesis in Chicken Liver. Int. J. Mol. Sci. 2020, 21, 1624. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Li, F.; Nie, L. Integrating multiple ‘omics’ analysis for microbial biology: Application and methodologies. Microbiology 2010, 156, 287–301. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Xiao, Q.; Gilbert, E.R.; Cui, Z.; Zhao, X.; Wang, Y.; Yin, H.; Li, D.; Zhang, H.; Zhu, Q. Whole-transcriptome analysis of atrophic ovaries in broody chickens reveals regulatory pathways associated with proliferation and apoptosis. Sci. Rep. 2018, 8, 7231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clavero, S.; Martínez, M.A.; Pérez, B.; Pérez-Cerdá, C.; Ugarte, M.; Desviat, L.R. Functional characterization of PCCA mutations causing propionic acidemia. BBA 2002, 1588, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zheng, X.; Feng, M.; Mo, Z.; Shan, Y.; Wang, Y.; Jin, J. Upregulated MMP28 in Hepatocellular Carcinoma Promotes Metastasis via Notch3 Signaling and Predicts Unfavorable Prognosis. Int. J. Biol. Sci. 2019, 15, 812–825. [Google Scholar] [CrossRef] [Green Version]
- Takada, Y.K.; Yu, J.; Shimoda, M.; Takada, Y. Integrin Binding to the Trimeric Interface of CD40L Plays a Critical Role in CD40/CD40L Signaling. J. Immunol. 2019, 203, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Nam, J.; Li, Y.; Kim, S.; Cho, S.H.; Cho, Y.S.; Choi, S.Y.; Choi, J.; Han, K.; Kim, Y.; et al. Regulation of dendritic spines, spatial memory, and embryonic development by the TANC family of PSD-95-interacting proteins. J. Neurosci. 2010, 30, 15102–15112. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Lei, Z.; Jiangjiu, L. Expression profiles of long noncoding RNAs in rat myocardial hypertrophy induced by pressure overload. J. Shandong Univ. Health Sci. 2015, 5, 21–26. [Google Scholar]







| Hormones | Infertile | Fertile | Negative Control |
|---|---|---|---|
| E2 (ng/L) | 72.7 ± 3.71 a | 75.88 ± 4.92 ab | 45.14 ± 5.87 c |
| T (ng/L) | 3531.95 ± 214.42 a | 3222.02 ± 282.10 a | 3599.93 ± 420.06 a |
| LH (ng/L) | 927.77 ± 126.99 a | 945.12 ± 151.38 a | 776.88 ± 204.02 a |
| FSH (U/L) | 26.22 ± 1.20 b | 28.78 ± 2.57 bc | 32.68 ± 3.48 a |
| VTG (μg/L) | 596.57 ± 59.56 b | 1564.13 ± 199.17 a | 298.43 ± 15.18 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mang, Q.; Hou, J.; Han, T.; Wang, G.; Wang, Y.; Liu, Y.; Ren, Y.; Zhao, Y.; He, Z.; Zhang, X. The Effect of Infertility on the Liver Structure, Endocrinology, and Gene Network in Japanese Flounder. Animals 2021, 11, 936. https://doi.org/10.3390/ani11040936
Mang Q, Hou J, Han T, Wang G, Wang Y, Liu Y, Ren Y, Zhao Y, He Z, Zhang X. The Effect of Infertility on the Liver Structure, Endocrinology, and Gene Network in Japanese Flounder. Animals. 2021; 11(4):936. https://doi.org/10.3390/ani11040936
Chicago/Turabian StyleMang, Qi, Jilun Hou, Tian Han, Guixing Wang, Yufen Wang, Yufeng Liu, Yuqin Ren, Yaxian Zhao, Zhongwei He, and Xiaoyan Zhang. 2021. "The Effect of Infertility on the Liver Structure, Endocrinology, and Gene Network in Japanese Flounder" Animals 11, no. 4: 936. https://doi.org/10.3390/ani11040936
APA StyleMang, Q., Hou, J., Han, T., Wang, G., Wang, Y., Liu, Y., Ren, Y., Zhao, Y., He, Z., & Zhang, X. (2021). The Effect of Infertility on the Liver Structure, Endocrinology, and Gene Network in Japanese Flounder. Animals, 11(4), 936. https://doi.org/10.3390/ani11040936





