Oryzias curvinotus in Sanya Does Not Contain the Male Sex-Determining Gene dmy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Collection of Fish and Samples
2.3. Morphological Traits and Molecular Species Identification
2.4. Genome Re-Sequencing
2.5. RNA Isolation, cDNA Library Preparation, and Sequencing
2.6. De Novo Assembly and Functional Annotation
2.7. Differential Gene Expression Analysis and Quantitative Real-Time PCR (qPCR) Validation
3. Results
3.1. Morphological and Molecular Analyses of SY-Medaka
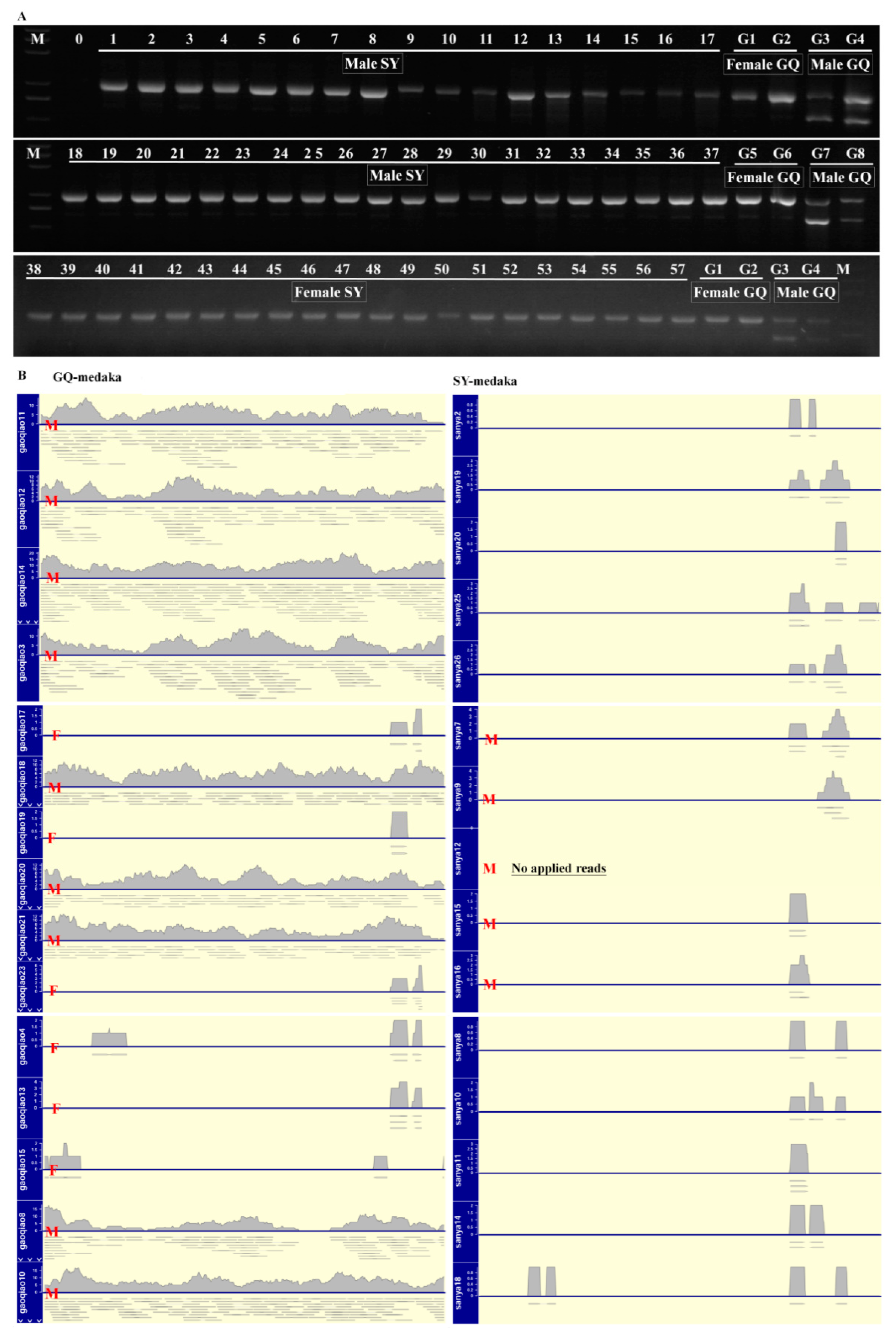
3.2. Genetic Sex Identification of SY-Medaka
3.3. RNA-Seq Analysis, Unigene Annotation, and dmy Detection
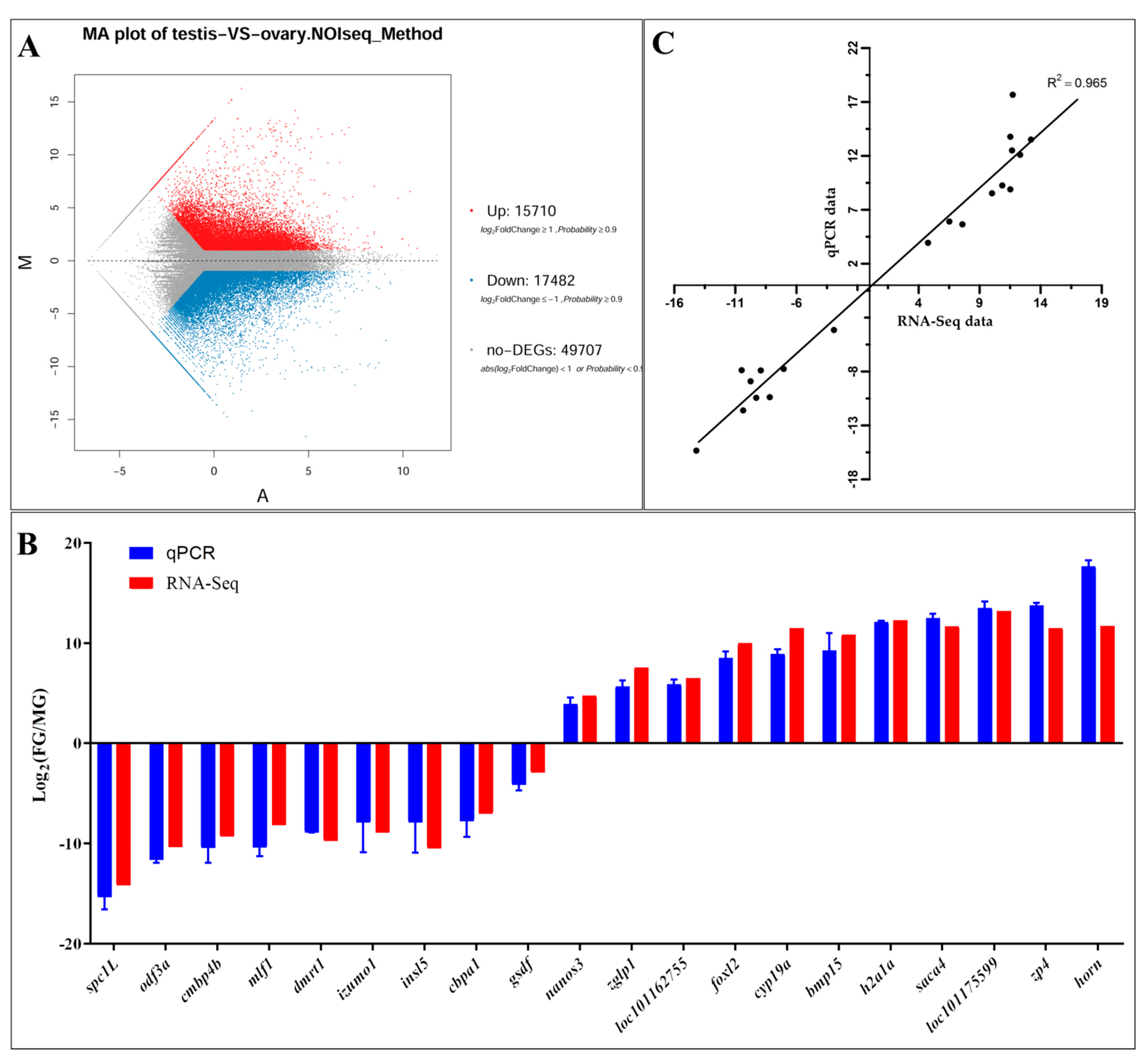
3.4. Differentially Expressed Gene Identification and qPCR Validation
4. Discussion
4.1. SY-Medaka Belongs to O. curvinotus.
4.2. SY-Medaka Does Not Contain dmy
4.3. Genes Related to Sex Determination and Gonadal Development in SY-Medaka
4.4. Lack of dmy in the SY-Medaka Genome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hayakawa, H.; Le, Q.D.; Kinoshita, M.; Takehana, Y.; Sakuma, K.; Takeshima, H.; Kojima, S.; Naruse, K.; Inoue, K. Genetic similarity of the Hainan medaka populations collected from hyper- and hypo-osmotic environments in northern Vietnam. Ocean. Sci. J. 2015, 50, 231–235. [Google Scholar] [CrossRef]
- Wang, Z.D.; Long, S.S.; Liao, J.; Huang, C.Q.; Zhang, H.R.; Huang, S.K.; Zhang, Y.P.; Liu, L.; Guo, Y.S. Complete mitogenome of Hainan medaka Oryzias curvinotus (Teleostei: Beloniformes and transcriptional differences between male and female liver. Mitochondrial DNA B 2017, 2, 157–158. [Google Scholar] [CrossRef]
- Matsuda, M.; Nagahama, Y.; Shinomiya, A.; Sato, T.; Matsuda, C.; Kobayashi, T.; Morrey, C.E.; Shibata, N.; Asakawa, S.; Shimizu, N.; et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002, 417, 559–563. [Google Scholar] [CrossRef]
- Matsuda, M.; Nagahama, Y.; Kobayashi, T.; Matsuda, C.; Hamaguchi, S.; Sakaizumi, M. The sex determining gene of medaka: A Y-specific DM domain gene (DMY is required for male development. Fish Physiol. Biochem. 2003, 28, 135–139. [Google Scholar] [CrossRef]
- Matsuda, M.; Sato, T.; Toyazaki, Y.; Nagahama, Y.; Hamaguchi, S.; Sakaizumi, M. Oryzias curvinotus has DMY, a gene that is required for male development in the medaka, O. latipes. Zool. Sci. 2003, 20, 159–161. [Google Scholar] [CrossRef]
- Kato, M.; Takehana, Y.; Sakaizumi, M.; Hamaguchi, S. A sex-determining region on the Y chromosome controls the sex-reversal ratio in interspecific hybrids between Oryzias curvinotus females and Oryzias latipes males. Heredity 2010, 104, 191–195. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yano, A.; Guyomard, R.; Nicol, B.; Jouanno, E.; Quillet, E.; Klopp, C.; Bouchez, O.; Fostier, A.; Guiguen, Y. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 2012, 22, 1423–1428. [Google Scholar] [CrossRef]
- Kamiya, T.; Kai, W.; Tasumi, S.; Oka, A.; Matsunaga, T.; Mizuno, N.; Fujita, M.; Suetake, H.; Suzuki, S.; Hosoya, S.; et al. A trans-species missense SNP in amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (Fugu). PLoS Genet. 2012, 8, e1002798. [Google Scholar] [CrossRef]
- Hattori, R.S.; Strüssmann, C.A. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. P. Natl. Acad. Sci. 2012, 109, 2955. [Google Scholar] [CrossRef] [PubMed]
- Myosho, T.; Otake, H.; Masuyama, H.; Matsuda, M.; Kuroki, Y.; Fujiyama, A.; Naruse, K.; Hamaguchi, S.; Sakaizumi, M. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 2012, 191, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.K.; Liu, Y.; Wang, W.W.; Wang, Q.; Zhang, N.; Lin, F.; Wang, N.; Shao, C.W.; Dong, Z.D.; Li, Y.Z.; et al. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 2017, 7, 42213. [Google Scholar] [CrossRef]
- Chen, S.L.; Zhang, G.J.; Shao, C.W.; Huang, Q.F.; Liu, G.; Zhang, P.; Song, W.T.; An, N.; Chalopin, D.; Volff, J.N.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Tao, W.J.; Chen, J.L.; Tan, D.J.; Yang, J.; Sun, L.N.; Wei, J.; Conte, M.A.; Kocher, T.D.; Wang, D.S. Transcriptome display during tilapia sex determination and differentiation as revealed by RNA-Seq analysis. BMC Genom. 2018, 19, 363. [Google Scholar] [CrossRef] [PubMed]
- He, F.X.; Jiang, D.N.; Huang, Y.Q.; Mustapha, U.F.; Yang, W.; Cui, X.F.; Tian, C.X.; Chen, H.P.; Shi, H.J.; Deng, S.P.; et al. Comparative transcriptome analysis of male and female gonads reveals sex-biased genes in spotted scat (Scatophagus argus). Fish Physiol. Biochem. 2019, 45, 1963–1980. [Google Scholar] [CrossRef]
- Tian, C.X.; Liu, Z.Y.; Dong, Z.D.; Huang, Y.; Du, T.; Chen, H.P.; Jiang, D.N.; Deng, S.P.; Zhang, Y.L.; Wanida, S.; et al. Transcriptome analysis of male and female mature gonads of silver sillago (Sillago sihama). Genes 2019, 10, 129. [Google Scholar] [CrossRef]
- Cadrin, S.X. Advances in morphometric identification of fishery stocks. Rev. Fish Biol. Fisher. 2000, 10, 91–112. [Google Scholar] [CrossRef]
- Frederich, B.; Liu, S.Y.V.; Dai, C.F. Morphological and genetic divergences in a coral reef damselfish, Pomacentrus coelestis. Evol biol. 2012, 39, 359–370. [Google Scholar] [CrossRef]
- Liu, X.H.; Song, N.; Liu, H.Y.; Li, Q.H.; Chen, Z.Y.; Yin, L.N.; Wang, Y.P.; Gao, T.X. Preliminary analysis on morphological characteristics of 5 Collichthys lucidus geographical populations. Trans. Oceanol. Limnol. 2015, 2, 59–67. [Google Scholar] [CrossRef]
- Wang, W.; Ma, C.Y.; Chen, W.; Jin, Z.W.; Zhao, M.; Zhang, F.Y.; Liu, Z.Q.; Ma, L.B. Population genetic diversity of mud crab (Scylla paramamosain from southeast coastal regions of China based on mitochondrial COI gene sequence. Gene 2020, 751, 144763. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.P.; Jamsari, A.F.J.; Muchlisin, Z.A.; Azizah, M.N.S. Mitochondrial genetic variation and population structure of the striped snakehead, Channa striata in Malaysia and Sumatra, Indonesia. Biochem. Syst. Ecol. 2015, 60, 99–105. [Google Scholar] [CrossRef]
- Zhu, S.R.; Fu, J.J.; Wang, Q.; Li, J.L. Identification of Channa species using the partial cytochrome c oxidase subunit I (COI gene as a DNA barcoding marker. Biochem. Syst. Ecol. 2013, 51, 117–122. [Google Scholar] [CrossRef]
- Zhang, F.F.; Jiang, H.C.; Jin, J.J.; Qiu, Y.P.; Chen, G.Z. Characteristic re-description of ricefish Oryzias curvinotus from Guangdong, China. Sichuan J. Zool. 2017, 36, 564–571. [Google Scholar] [CrossRef]
- Dong, Z.D.; Long, S.S.; Huang, C.Q.; Huang, S.K.; Zhang, N.; Ying, Z.Y.; Guo, Y.S.; Wang, Z.D. A method for rapid identification of genetic sex of Oryzias curvinotus. J. Guangdong Ocean Univ. 2018, 38, 25–29. [Google Scholar] [CrossRef]
- Lin, Y.S.; Wang, B.; Wang, N.H.; Ouyang, G.; Cao, H. Transcriptome analysis of rare minnow (Gobiocypris rarus infected by the grass carp reovirus. Fish Shellfish Immunol. 2019, 89, 337–344. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 16. [Google Scholar] [CrossRef]
- Tarazona, S.; Garcia-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef]
- Dong, Z.D.; Chen, P.S.; Zhang, N.; Huang, S.K.; Zhang, H.R.; Wang, S.R.; Li, X.Y.; Guo, Y.S.; Wang, Z.D. Evaluation of reference genes for quantitative real-time PCR analysis of gene expression in Hainan medaka (Oryzias curvinotus). Gene Rep. 2019, 14, 94–99. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)). Method 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mayr, E.; Linsley, E.G.; Usinger, R. Methods and Principles of Systematic Zoology; McGraw Hill: New York, NY, USA, 1953; pp. 22–39. [Google Scholar]
- Takehana, Y.; Naruse, K.; Sakaizumi, M. Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol. Phylogenet Evol. 2005, 36, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Mokodongan, D.F.; Yamahira, K. Origin and intra-island diversification of Sulawesi endemic Adrianichthyidae. Mol. Phylogenet Evol. 2015, 93, 150–160. [Google Scholar] [CrossRef]
- Hellberg, R.S.; Kawalek, M.D.; Van, K.T.; Shen, Y.L.; Williams-Hill, D.M. Comparison of DNA Extraction and PCR Setup Methods for Use in High-Throughput DNA Barcoding of Fish Species. Food Anal. Methods 2014, 7, 1950–1959. [Google Scholar] [CrossRef]
- Leon, H.; Julia, S. The natural history of model organisms: The untapped potential of medaka and its wild relatives. eLife 2019, 8, e46994. [Google Scholar] [CrossRef]
- Liu, H.R.; Zhang, H.; Pan, X.L.; Xu, M.; Huang, J.; He, M.X. A high density genetic map by whole-genome resequencing for QTL fine-mapping and dissecting candidate genes for growth or sex traits in the pearl oyster (Pinctada fucata martensii). Aquaculture 2020, 519, 734839. [Google Scholar] [CrossRef]
- Lin, Q.H.; Mei, J.; Li, Z.; Zhang, X.M.; Zhou, L.; Gui, J.F. Distinct and cooperative roles of amh and dmrt1 in self-renewal and differentiation of male germ cells in zebrafish. Genetics. 2017, 207, 1007–1022. [Google Scholar] [CrossRef]
- Nakamura, S.; Watakabe, I.; Nishimura, T.; Picard, J.Y.; Toyoda, A.; Taniguchi, Y.; di Clemente, N.; Tanaka, M. Hyperproliferation of mitotically active germ cells due to defective anti-Mullerian hormone signaling mediates sex reversal in medaka. Development 2012, 139, 2283–2287. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.N.; Yang, H.H.; Li, M.H.; Shi, H.J.; Zhang, X.B.; Wang, D.S. gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the nile tilapia. Mol. Reprod. Dev. 2016, 83, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guan, G.J.; Li, M.Y.; Zhu, F.; Hong, Y.H. Autosomal gsdf acts as a male sex initiator in the fish medaka. Sci. Rep. 2016, 6, 19738. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Watakabe, I.; Nishimura, T.; Toyoda, A.; Taniguchi, Y.; Tanaka, M. Analysis of medaka sox9 orthologue reveals a conserved role in germ cell maintenance. PLoS ONE 2012, 7, 12. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, J.; Li, Y.; Zhou, Z.L.; Gui, J.F. Sequential, divergent, and cooperative requirements of foxl2a and foxl2b in ovary development and maintenance of zebrafish. Genetics 2017, 205, 1551–1572. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Y.; Wang, Y.J.; Wang, M.Y.; Liu, Y.X.; Cheng, J.; Zhang, Q.Q. Growth differentiation factor 9 (gdf9) and bone morphogenetic protein 15 (bmp15) are potential intraovarian regulators of steroidogenesis in Japanese flounder (Paralichthys olivaceus). Gen. Comp. Endocrinol. 2020, 297, 113547. [Google Scholar] [CrossRef] [PubMed]
- Dranow, D.B.; Hu, K.; Bird, A.M.; Lawry, S.T.; Adams, M.T.; Sanchez, A.; Amatruda, J.F.; Draper, B.W. Bmp15 is an oocyte-produced signal required for maintenance of the adult female sexual phenotype in zebrafish. PLoS Genet. 2016, 12, 24. [Google Scholar] [CrossRef]
- Wu, T.L.; Cheng, Y.Y.; Liu, Z.L.; Tao, W.J.; Zheng, S.Q.; Wang, D.S. Bioinformatic analyses of zona pellucida genes in vertebrates and their expression in Nile tilapia. Fish Physiol. Biochem. 2018, 44, 435–449. [Google Scholar] [CrossRef]
- Dong, Z.D.; Zhang, N.; Liu, Y.; Xu, W.T.; Cui, Z.K.; Shao, C.W.; Chen, S.L. Expression analysis and characterization of zglp1 in the Chinese tongue sole (Cynoglossus semilaevis). Gene 2019, 683, 72–79. [Google Scholar] [CrossRef]
- Miao, L.Y.; Yuan, Y.; Cheng, F.; Fang, J.S.; Zhou, F.; Ma, W.R.; Jiang, Y.; Huang, X.H.; Wang, Y.C.; Shan, L.J.; et al. Translation repression by maternal RNA binding protein Zar1 is essential for early oogenesis in zebrafish. Development 2017, 144, 128–138. [Google Scholar] [CrossRef]
- Corina, H.; Walter, S.; Astrid, B. Genetics of sexual development: An evolutionary playground for fish. Genetics 2014, 196, 579–591. [Google Scholar] [CrossRef]
- Stelkens, R.B.; Wedekind, C. Environmental sex reversal, Trojan sex genes, and sex ratio adjustment: Conditions and population consequences. Mol. Ecol. 2010, 19, 627–646. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Zhang, Y.; Sarida, M. Coexistence of genotypic and temperature-dependent sex determination in pejerrey Odontesthes bonariensis. PLoS ONE 2014, 9, e102574. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Endo, T.; Yamahira, K.; Hamaguchi, S.; Sakaizumi, M. Induction of female-to-male sex reversal by high temperature treatment in medaka, Oryzias latipes. Zool. Sci. 2005, 22, 985–988. [Google Scholar] [CrossRef]
- Shao, C.W.; Li, Q.Y.; Chen, S.L.; Zhang, P.; Lian, J.M.; Hu, Q.M.; Sun, B.; Jin, L.J.; Liu, S.S.; Wang, Z.J.; et al. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 2014, 24, 604–615. [Google Scholar] [CrossRef]
- Desprez, D.; Melard, C. Effect of ambient water temperature on sex determinism in the blue tilapia Oreochromis Aureus. Aquaculture 1998, 162, 79–84. [Google Scholar] [CrossRef]
- Myosho, T.; Takehana, Y.; Hamaguchi, S.; Sakaizumi, M. Turnover of sex chromosomes in celebensis group medaka fishes. G3-Genes Genom. Genet. 2015, 5, 2685–2691. [Google Scholar] [CrossRef] [PubMed]
- Takehana, Y.; Matsuda, M.; Myosho, T.; Suster, M.L.; Kawakami, K.; Shin-I, T.; Kohara, Y.; Kuroki, Y.; Toyoda, A.; Fujiyama, A.; et al. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 2014, 5, 4157. [Google Scholar] [CrossRef] [PubMed]
- Herpin, A.; Braasch, I.; Kraeussling, M.; Schmidt, C.; Thoma, E.C.; Nakamura, S.; Tanaka, M.; Schartl, M. Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements. PLoS Genet. 2010, 6, 15. [Google Scholar] [CrossRef]
- Matsuda, M.; Sakaizumi, M. Evolution of the sex-determining gene in the teleostean genus Oryzias. Gen. Comp. Endocrinol. 2016, 239, 80–88. [Google Scholar] [CrossRef] [PubMed]


| Countable Traits | GQ-Medaka | SY-Medaka | Variable Coefficient |
|---|---|---|---|
| dorsal fin ray counts | 6.02 ± 0.15 | 6.04 ± 0.19 | 0.05 |
| anal fin ray counts | 19.49 ± 0.87 | 19.71 ± 0.80 | 0.13 |
| pectoral fin ray counts | 8.09 ± 0.29 | 8.12 ± 0.32 | 0.05 |
| caudal fin ray counts | 18.51 ± 0.69 b | 19.31 ± 1.02 a | 0.47 |
| ventral fin ray counts | 6.00 ± 0.00 | 6.00 ± 0.00 | |
| measurable traits | |||
| Body weight/g | 0.203 ± 0.06 | 0.204 ± 0.04 | 0.01 |
| Total length/mm | 27.71 ± 2.25 a | 27.59 ± 1.83 b | 0.03 |
| Head length/mm | 5.20 ± 0.62 b | 5.44 ± 0.50 a | 0.2 |
| Snount length/mm | 1.21 ± 0.25 | 1.29 ± 0.22 | 0.17 |
| Eye orbit diameter/mm | 2.06 ± 0.27 | 2.00 ± 0.22 | 0.12 |
| Maximum depth of body/mm | 5.01 ± 0.72 | 5.12 ± 0.44 | 0.09 |
| Length of caudal fin/mm | 4.32 ± 0.35 | 4.21 ± 0.31 | 0.17 |
| Tips of snout to anus/mm | 12.22 ± 0.94 | 12.04 ± 0.80 | 0.1 |
| Tip of snout to dorsal fin/mm | 13.14 ± 0.98 a | 12.63 ± 0.92 b | 0.27 |
| Caudal peduncle length/mm | 4.19 ± 0.33 | 4.17 ± 0.23 | 0.04 |
| Caudal peduncle depth/mm | 2.09 ± 0.25 b | 2.24 ± 0.18 a | 0.35 |
| Length of dorsal fin/mm | 3.30 ± 0.67 b | 3.68 ± 0.67 a | 0.28 |
| Length of base of anal fin/mm | 2.78 ± 0.35 b | 2.96 ± 0.41 a | 0.23 |
| Length of pectoral fin/mm | 3.90 ± 0.41 b | 4.36 ± 0.37 a | 0.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Z.; Li, X.; Yao, Z.; Wang, C.; Guo, Y.; Wang, Q.; Shao, C.; Wang, Z. Oryzias curvinotus in Sanya Does Not Contain the Male Sex-Determining Gene dmy. Animals 2021, 11, 1327. https://doi.org/10.3390/ani11051327
Dong Z, Li X, Yao Z, Wang C, Guo Y, Wang Q, Shao C, Wang Z. Oryzias curvinotus in Sanya Does Not Contain the Male Sex-Determining Gene dmy. Animals. 2021; 11(5):1327. https://doi.org/10.3390/ani11051327
Chicago/Turabian StyleDong, Zhongdian, Xueyou Li, Zebin Yao, Chun Wang, Yusong Guo, Qian Wang, Changwei Shao, and Zhongduo Wang. 2021. "Oryzias curvinotus in Sanya Does Not Contain the Male Sex-Determining Gene dmy" Animals 11, no. 5: 1327. https://doi.org/10.3390/ani11051327
APA StyleDong, Z., Li, X., Yao, Z., Wang, C., Guo, Y., Wang, Q., Shao, C., & Wang, Z. (2021). Oryzias curvinotus in Sanya Does Not Contain the Male Sex-Determining Gene dmy. Animals, 11(5), 1327. https://doi.org/10.3390/ani11051327






