Seasonal Variation in Gut Microbiota Related to Diet in Fejervarya limnocharis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Diet Analyses
2.3. DNA Extraction and Sequencing
2.4. Operational Taxonomic Unit (OTU) Cluster and Species Annotation
2.5. Data Analysis
2.6. Function Prediction
3. Results
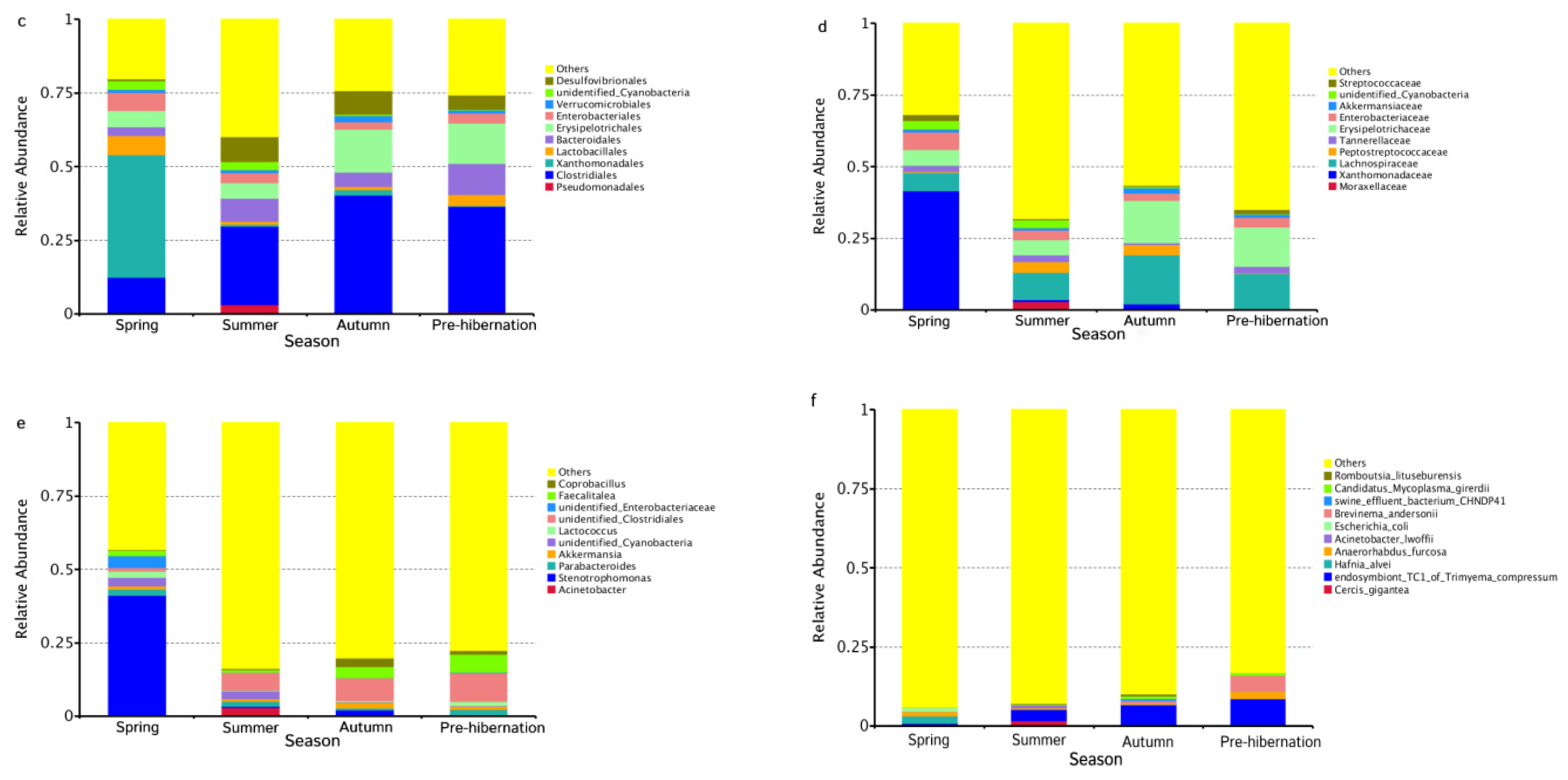
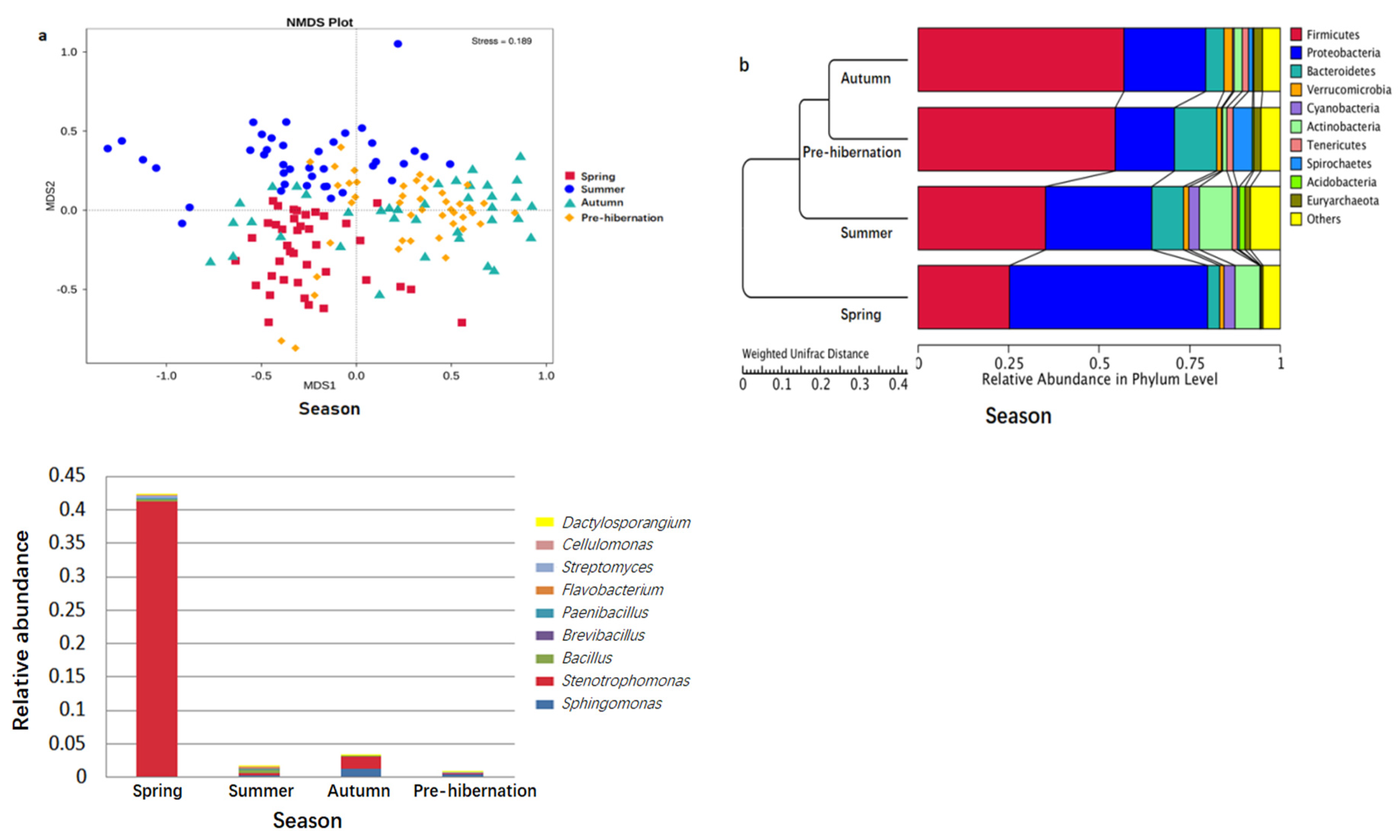
3.1. Gut Microbiota Composition of F. limnocharis
3.2. Seasonal Changes of Intestinal Microbiota in Frogs
3.3. Variations of Function Prediction in Different Seasons
3.4. Dietary Changes
3.5. Explanatory Factors of Intestinal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McNab, B.K. The Physiological Ecology of Vertebrates: A View from Energetics; Cornell University Press: Ithaca, NY, USA, 2002. [Google Scholar]
- Walk, S.T.; Blum, A.M.; Ewing, S.A.S.; Weinstock, J.V.; Young, V.B. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm. Bowel Dis. 2010, 16, 1841–1849. [Google Scholar] [CrossRef]
- Linnenbrink, M.; Wang, J.; Hardouin, E.A.; Künzel, S.; Metzler, D.; Baines, J.F. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol. Ecol. 2013, 22, 1904–1916. [Google Scholar] [CrossRef]
- Pryor, G.S. Anaerobic bacteria isolated from the gastrointestinal tracts of bullfrog tadpoles (Rana catesbeiana). Herpetol. Conserv. Biol. 2008, 3, 176–181. [Google Scholar]
- Huang, C.H.; Yu, X.; Liao, W.B. The expensive-tissue hypothesis in vertebrates: Gut microbiota effect, a review. Int. J. Mol. Sci. 2018, 19, 1792. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227. [Google Scholar] [CrossRef]
- Zhang, Y. The Community Structure and Functional Genes of the Chicken Gut Microorganism and the Chicken Growth. Ph.D. Thesis, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China, 2018. [Google Scholar]
- Rana, S.W.; Kumar, A.; Walia, S.K.; Berven, K.; Cumper, K.; Walia, S.K. Isolation of Tn1546-like elements in vancomycin-resistant Enterococcus faecium isolated from wood frogs: An emerging risk for zoonotic bacterial infections to humans. J. Appl. Microbiol. 2011, 110, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Sanz, Y.; De Palma, G.; Laparra, M. Unraveling the ties between celiac disease and intestinal microbiota. Int. Rev. Immunol. 2011, 30, 207–218. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.M.; Wu, Y.P.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, Y.; Gao, J.; Li, X. Effects of octylphenol exposure on the lipid metabolism and microbiome of the intestinal tract of Rana chensinensis tadpole by RNAseq and 16s amplicon sequencing. Ecotoxicol. Environ. Saf. 2020, 197, 110650. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.K.; Kelly, S.A.; Legge, R.; Ma, F.; Low, S.J.; Kim, J.; Zhang, M.; Oh, P.L.; Nehrenberg, D.; Hua, K.; et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18933–18938. [Google Scholar] [CrossRef] [PubMed]
- Vangay, P.; Johnson, A.J.; Ward, T.L.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Hillmann, B.M.; Lucas, S.K.; Beura, L.K.; Thompson, E.A.; Till, L.M.; et al. US immigration westernizes the human gut microbiome. Cell 2018, 175, 962–972.e10. [Google Scholar] [CrossRef] [PubMed]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013, 7, 1344. [Google Scholar] [CrossRef]
- Maurice, C.F.; Knowles, S.C.L.; Ladau, J.; Pollard, K.S.; Fenton, A.; Pedersen, A.B.; Turnbaugh, P.J. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015, 9, 2423. [Google Scholar] [CrossRef]
- Bletz, M.C.; Goedbloed, D.J.; Sanchez, E.; Reinhardt, T.; Tebbe, C.C.; Bhuju, S.; Geffers, R.; Jarek, M.; Vences, M.; Steinfartz, S. Amphibian gut microbiota shifts differentially in community structure but converges on habitat-specific predicted functions. Nat. Commun. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Hamilton, M.J.; Staley, C.; Sadowsky, M.J.; Sorensen, P.W. Environment shapes the fecal microbiome of invasive carp species. Microbiome 2016, 4, 44. [Google Scholar] [CrossRef]
- Kohl, K.D.; Yahn, J. Effects of environmental temperature on the gut microbial communities of tadpoles. Environ. Microbiol. 2016, 18, 1561–1565. [Google Scholar] [CrossRef]
- Tong, Q.; Cui, L.Y.; Hu, Z.F.; Du, X.P.; Abid, H.M.; Wang, H.B. Environmental and host factors shaping the gut microbiota diversity of brown frog Rana dybowskii. Sci. Total Environ. 2020, 741, 140142. [Google Scholar] [CrossRef]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; González, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974. [Google Scholar] [CrossRef]
- Bolnick, D.I.; Snowberg, L.K.; Hirsch, P.E.; Lauber, C.L.; Knight, R.; Caporaso, J.G.; Svanbäck, R. Individuals’ diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch). Ecol. Lett. 2014, 17, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Hicks, A.L.; Lee, K.J.; Couto-Rodriguez, M.; Patel, J.; Sinha, R.; Guo, C.; Olson, S.H.; Seimon, A.; Seimon, T.A.; Ondzie, A.U.; et al. Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat. Commun. 2018, 9, 1786. [Google Scholar] [CrossRef] [PubMed]
- Weng, F.C.H.; Yang, Y.J.; Wang, D. Functional analysis for gut microbes of the brown tree frog Polypedates megacephalus in artificial hibernation. BMC Genom. 2016, 17, 1024. [Google Scholar] [CrossRef]
- Smits, S.A.; Leach, J.; Sonnenburg, E.D.; Gonzalez, C.G.; Lichtman, J.S.; Knight, P.; Manjurano, A.; Changalucha, J.; Elias, J.E.; Dominguez-Bello, M.G.; et al. Seasonal cycling in the gut microbiome of the Hadza huntergatherers of Tanzania. Science 2017, 357, 802–806. [Google Scholar] [CrossRef] [PubMed]
- Laparra, J.M.; Sanz, Y. Interactions of gut microbiota with functional food components and nutraceuticals. Pharmacol. Res. 2010, 61, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef]
- Bolnick, D.I.; Lau, O.L. Predictable patterns of disruptive selection in stickleback in postglacial lakes. Am. Nat. 2008, 172, 1–11. [Google Scholar] [CrossRef]
- Liao, W.B.; Lu, X.; Jehle, R. Altitudinal variation in maternal investment and trade-offs between egg size and clutch size in the Andrew’s toad. J. Zool. 2014, 293, 84–91. [Google Scholar] [CrossRef]
- Mai, C.L.; Liao, W.B.; Lüpold, S.; Kotrschal, A. Relative brain size is predicted by the intensity of intrasexual competition in frogs. Am. Nat. 2020, 196, 169–179. [Google Scholar] [CrossRef]
- Chang, C.W.; Huang, B.H.; Lin, S.M.; Huang, C.L.; Liao, P.C. Changes of diet and dominant intestinal microbes in farmland frogs. BMC Microbiol. 2016, 16, 33. [Google Scholar] [CrossRef] [PubMed]
- Mai, C.L.; Yu, J.P.; Liao, W.B. Ecological and geographical reasons for the variation of digestive tract length in Anurans. Asian Herpetol. Res. 2019, 10, 246. [Google Scholar]
- Mai, C.; Huang, J.; Liao, Q.; Liao, W.B.; Zhang, L.X. Altitudinal variation in digestive tract length in Feirana quadranus. Asian Herpetol. Res. 2019, 10, 183–189. [Google Scholar]
- Tong, Q. Diversity and Influencing Factors of Gut Microbiota in Rana dybowskii. Ph.D. Thesis, Northeast Forestry University, Harbin, China, 2019. [Google Scholar]
- Stevens, C.E. Digestive System of Amphibians, Reptiles and Birds; John Wiley & Sons, Ltd.: Chichester, West Sussex, UK, 2001. [Google Scholar]
- Liao, W.B.; Liu, W.C.; Merilä, J. Andrew meets Rensch: Sexual size dimorphism and the inverse of Rensch’s rule in Andrew’s toad (Bufo andrewsi). Oecologia 2015, 177, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Magurran, A.E. Ecological Diversity and Its Measurement; Croom Helm, Ltd.: London, UK, 1988. [Google Scholar]
- Colwell, R.K.; Fuluyma, D.J. Measurement of niche breath and overlap. Ecology 1971, 52, 567–576. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Rui, J.; Li, Y.; Tang, N.; Zhan, S.P.; Jiang, J.P.; Li, X.Z. Ambient temperature alters body size and gut microbiota of Xenopus tropicalis. Sci. China Life Sci. 2020, 63, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.H.; Chang, C.W.; Huang, C.W.; Gao, J.; Liao, P.C. Composition and functional specialists of the gut microbiota of frogs reflect habitat differences and agricultural activity. Front. Microbial. 2018, 8, 2670. [Google Scholar] [CrossRef]
- Shu, Y.; Hong, P.; Tang, D.; Qing, H.; Donde, O.O.; Wang, H.; Xiao, B.D.; Wu, H.L. Comparison of intestinal microbes in female and male Chinese concave-eared frogs (Odorrana tormota) and effect of nematode infection on gut bacterial communities. MicrobiologyOpen 2019, 8, e00749. [Google Scholar] [CrossRef]
- Tong, Q.; Zhang, J. Effects of captivity and season on the gut microbiota of the brown frog (Rana dybowskii). Front. Microbiol. 2019, 10, 1912. [Google Scholar]
- Tong, Q.; Hu, Z.F.; Du, X.P.; Bie, J.; Wang, H.B. Effects of seasonal hibernation on the similarities between the skin microbiota and gut microbiota of an amphibian (Rana dybowskii). Microb. Ecol. 2019, 79, 898–909. [Google Scholar] [CrossRef]
- Antwis, R.E.; Haworth, R.L.; Engelmoer, D.J.; Ogilvy, V.; Fidgett, A.L.; Preziosi, R.F. Ex situ diet influences the bacterial community associated with the skin of red-eyed tree frogs (Agalychnis callidryas). PLoS ONE 2014, 9, e85563. [Google Scholar] [CrossRef] [PubMed]
- Henneron, L.; Bernard, L.; Hedde, M.; Pelosi, C.; Villenave, C.; Chenu, C.; Bertrand, M.; Girardin, C.; Blanchart, E. Fourteen years of evidence for positive effects of conservation agriculture and organic farming on soil life. Agron. Sustain. Dev. 2015, 35, 169–181. [Google Scholar] [CrossRef]
- Rice, J.A. Does Gut Microbiota Affect the Diet Preference in Anurans? University of Southern Mississippi: Hattiesburg, MS, USA, 2015. [Google Scholar]
- Glorioso, J.C.; Amborski, R.L.; Amborski, G.F.; Culley, D.D. Microbiological studies on septicemic bullfrogs (Rana catesbeiana). Am. J. Vet. Res. 1974, 35, 1241–1245. [Google Scholar]
- Bresciano, J.C.; Salvador, C.A.; Paz-Y-Miño, C.; Parody-Merino, A.M.; Bosch, J.; Woodhams, D.C. Variation in the presence of anti-Batrachochytrium dendrobatidis bacteria of amphibians across life stages and elevations in Ecuador. Ecohealth 2015, 12, 310–319. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut Microbiota and Immune System Interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef]
- Macpherson, A.J.; McCoy, K.D.; Johansen, F.E.; Brandtzaeg, P. The immune geography of IgA induction and function. Nat. Rev. 2008, 1, 11–22. [Google Scholar] [CrossRef]
- Neish, A.S. Microbes in gastrointestinal health and disease. Gastroenterology 2009, 136, 65–80. [Google Scholar] [CrossRef]
- Russell, J.B.; Rychlik, J.L. Factors that alter rumen microbial ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef]
- Candela, M.; Biagi, E.; Maccaferri, S.; Turroni, S.; Brigidi, P. Intestinal microbiota is a plastic factor responding to environmental changes. Trends Microbiol. 2012, 20, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Alberdi, A.; Aizpurua, O.; Bohmann, K.; Zepeda-Mendoza, M.L.; Gilbert, M.T.P. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 2016, 31, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Macke, E.; Tasiemski, A.; Massol, F.; Callens, M.; Decaestecker, E. Life history and eco-evolutionary dynamics in light of the gut microbiota. Oikos 2017, 126, 508–531. [Google Scholar] [CrossRef]
- De La Cuesta-Zuluaga, J.; Corrales-Agudelo, V.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Body size phenotypes comprehensively assess cardiometabolic risk and refine the association between obesity and gut microbiota. Int. J. Obes. 2018, 42, 424–432. [Google Scholar] [CrossRef] [PubMed]





| Coefficients | Wald Chisq | df | p |
|---|---|---|---|
| Intercept | 1306.936 | 1 | <0.0001 |
| Diet diversity | 14.279 | 2 | 0.001 |
| Body mass | 1597.758 | 88 | 0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Liao, W. Seasonal Variation in Gut Microbiota Related to Diet in Fejervarya limnocharis. Animals 2021, 11, 1393. https://doi.org/10.3390/ani11051393
Huang C, Liao W. Seasonal Variation in Gut Microbiota Related to Diet in Fejervarya limnocharis. Animals. 2021; 11(5):1393. https://doi.org/10.3390/ani11051393
Chicago/Turabian StyleHuang, Chunhua, and Wenbo Liao. 2021. "Seasonal Variation in Gut Microbiota Related to Diet in Fejervarya limnocharis" Animals 11, no. 5: 1393. https://doi.org/10.3390/ani11051393
APA StyleHuang, C., & Liao, W. (2021). Seasonal Variation in Gut Microbiota Related to Diet in Fejervarya limnocharis. Animals, 11(5), 1393. https://doi.org/10.3390/ani11051393





