Systematic Analysis of Long Noncoding RNA and mRNA in Granulosa Cells during the Hen Ovulatory Cycle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Preparation and Sample Collection
2.2. RNA Extraction, Library Construction, and Sequencing
2.3. Transcriptome Annotation and Classification
2.4. Differential Expression Analysis
2.5. Short Time-Series Expression Miner Analysis
2.6. Functional Enrichment Analysis
2.7. Cell Culture and LH Treatment
2.8. Quantitative Real-Time PCR Validation
2.9. Statistical Analysis
3. Results
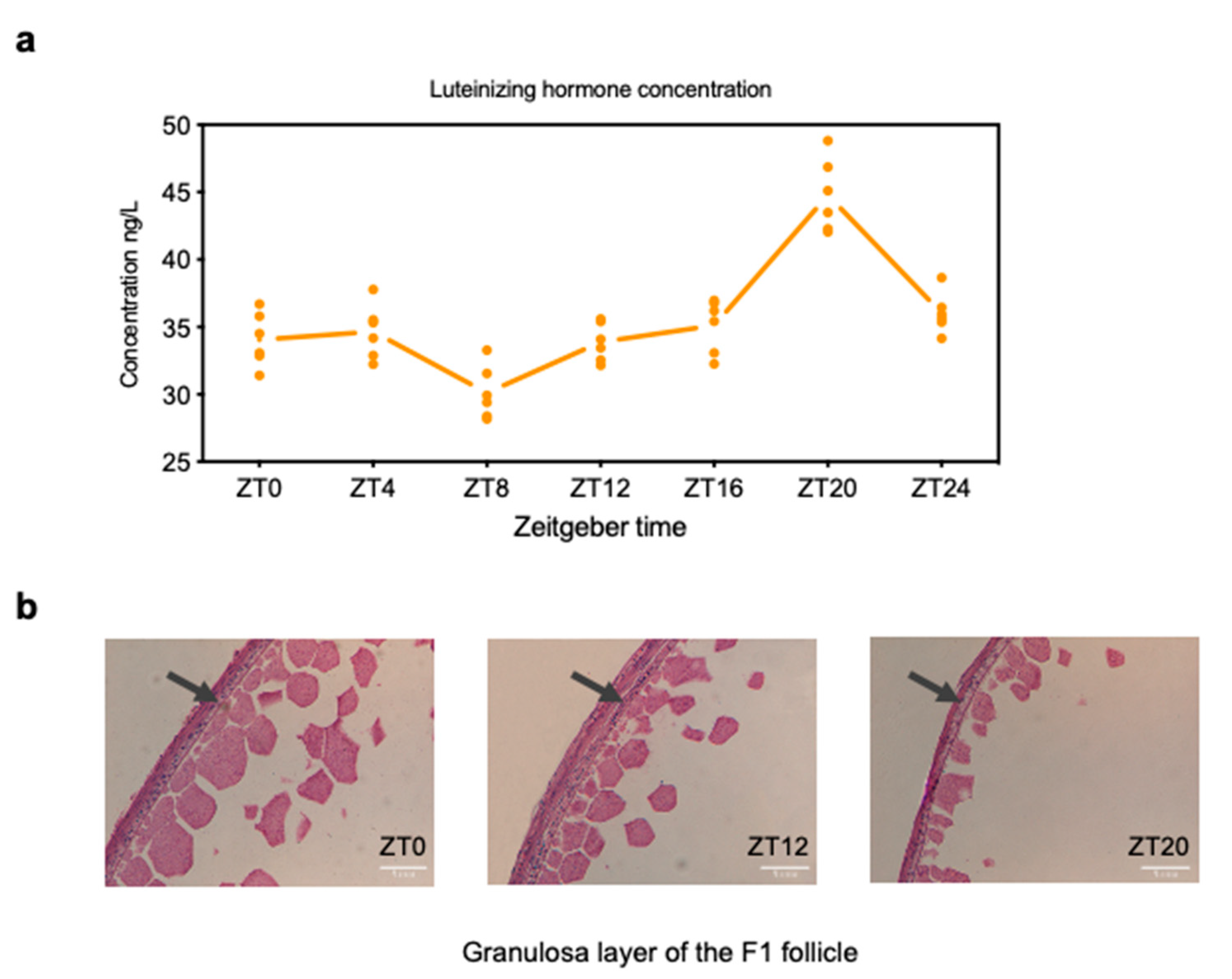
3.1. Changes in LH level and Histological Analyses of Chicken Follicle
3.2. Expression Pattern of lncRNAs and mRNAs
3.3. Functional Enrichment Analysis of Differentially Expressed mRNAs
3.4. Functional Enrichment Analysis of Differentially Expressed lncRNAs
3.5. Temporal Gene Expression Patterns of mRNAs and lncRNAs
3.6. Dynamic Expression of Follicular Steroid-Related Genes
3.7. Validation of mRNAs and lncRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| lncRNA | Long Non-coding RNA |
| LH | Luteinizing hormone |
| ZT | Zeitgeber time |
| ZT0 | The early stage of the ovulatory cycle at 9:00 a.m. |
| ZT12 | The middle stage of the ovulatory cycle at 9:00 p.m. |
| ZT20 | The LH surge stage at 5:00 a.m. |
| LH | Luteinizing Hormone |
| TPM | Transcripts Per Kilobase Million |
| STEM | Short time-series expression miner |
| DEGs | Differentially expressed genes |
| DELs | Differentially expressed lncRNAs |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DXM | Dexamethasone |
| qPCR | Quantitative real-time PCR |
| H&E | Hematoxylin and Eosin |
| t-SNE | t-Distributed stochastic neighbor embedding |
| GO | Gene Ontology |
| PDGFA | Platelet-Derived Growth Factor Subunit A |
| FASN | Fatty Acid Synthase |
| RGS16 | Regulator of G protein signaling 16 |
| EGFR | Epidermal Growth Factor Receptor |
| RGS2 | Regulator of G Protein Signaling 2 |
| STAR | Steroidogenic Acute Regulatory Protein |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Preisinger, R.; Flock, D.K. Genetic Changes in Layer Breeding: Historical Trends and Future Prospects. BSAP Occas. Publ. 2000, 27, 20–28. [Google Scholar] [CrossRef]
- Groothuis, T.G.G.; Schwabl, H. Hormone-Mediated Maternal Effects in Birds: Mechanisms Matter but What Do We Know of Them? Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 1647–1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, A.L. Reproduction in the Female. Sturkie’s Avian Physiol. 2015, 635–665. [Google Scholar] [CrossRef]
- Lovell, T.M.; Gladwell, R.T.; Groome, N.P.; Knight, P.G. Ovarian Follicle Development in the Laying Hen Is Accompanied by Divergent Changes in Inhibin A, Inhibin B, Activin A and Follistatin Production in Granulosa and Theca Layers. J. Endocrinol. 2003, 177, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, M.; Lee, D.; Panigone, S.; Horner, K.; Chen, R.; Theologis, A.; Lee, D.C.; Threadgill, D.W.; Conti, M. Luteinizing Hormone-Dependent Activation of the Epidermal Growth Factor Network Is Essential for Ovulation. Mol. Cell. Biol. 2007, 27, 1914–1924. [Google Scholar] [CrossRef] [Green Version]
- Laguë, P.C.; Van Tienhoven, A.; Cunningham, F.J. Concentrations of Estrogens, Progesterone and LH During the Ovulatory Cycle of the Laying Chicken [Gallus Domesticus]. Biol. Reprod. 1975, 12, 590–598. [Google Scholar] [CrossRef] [Green Version]
- Etches, R.J.; Schoch, J.P. A Mathematical Representation of the Ovulatory Cycle of the Domestic Hen. Br. Poult. Sci. 1984, 25, 65–76. [Google Scholar] [CrossRef]
- Nakao, N.; Yasuo, S.; Nishimura, A.; Yamamura, T.; Watanabe, T.; Anraku, T.; Okano, T.; Fukada, Y.; Sharp, P.J.; Ebihara, S.; et al. Circadian Clock Gene Regulation of Steroidogenic Acute Regulatory Protein Gene Expression in Preovulatory Ovarian Follicles. Endocrinology 2007, 148, 3031–3038. [Google Scholar] [CrossRef] [Green Version]
- Etches, R.J.; Petitte, J.N.; Anderson-Langmuir, C.E. Interrelationships between the Hypothalamus, Pituitary Gland, Ovary, Adrenal Gland, and the Open Period for LH Release in the Hen (Gallus Domesticus). J. Exp. Zool. 1984, 232, 501–511. [Google Scholar] [CrossRef]
- Sterneck, E.; Tessarollo, L.; Johnson, P.F. An Essential Role for C/EBPβ in Female Reproduction. Genes Dev. 1997, 11, 2153–2162. [Google Scholar] [CrossRef]
- Schuster, M.K.; Schmierer, B.; Shkumatava, A.; Kuchler, K. Activin A and Follicle-Stimulating Hormone Control Tight Junctions in Avian Granulosa Cells by Regulating Occludin Expression. Biol. Reprod. 2004, 70, 1493–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, J.G.; Parsons, T.F. Glycoprotein Hormones: Structure and Function. Annu. Rev. Biochem. 1981, 50, 465–495. [Google Scholar] [CrossRef] [PubMed]
- Doi, O.; Takai, T.; Nakamura, T.; Tanabe, Y. Changes in the Pituitary and Plasma LH, Plasma and Follicular Progesterone and Estradiol, and Plasma Testosterone and Estrone Concentrations during the Ovulatory Cycle of the Quail (Coturnix Coturnix Japonica). Gen. Comp. Endocrinol. 1980, 41, 156–163. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Q.; Liu, Z.; Guo, X.; Du, Y.; Yuan, Z.; Guo, M.; Kang, L.; Sun, Y.; Jiang, Y. Transcriptome Analysis on Single Small Yellow Follicles Reveals That Wnt4 Is Involved in Chicken Follicle Selection. Front. Endocrinol. 2017, 8, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Wang, Y.; Liu, Z.; Guo, X.; Sun, Y.; Kang, L.; Jiang, Y. Transcriptomic and Proteomic Analyses of Ovarian Follicles Reveal the Role of VLDLR in Chicken Follicle Selection. BMC Genom. 2020, 21, 486. [Google Scholar] [CrossRef]
- Luo, W.; Gu, L.; Li, J.; Gong, Y. Transcriptome Sequencing Revealed That Knocking down FOXL2 Affected Cell Proliferation, the Cell Cycle, and DNA Replication in Chicken Pre-Ovulatory Follicle Cells. PLoS ONE 2020, 15, e0234795. [Google Scholar] [CrossRef]
- Zhou, S.; Ma, Y.; Zhao, D.; Mi, Y.; Zhang, C. Transcriptome Profiling Analysis of Underlying Regulation of Growing Follicle Development in the Chicken. Poult. Sci. 2020, 99, 2861–2872. [Google Scholar] [CrossRef]
- Shen, M.; Wu, P.; Li, T.; Wu, P.; Chen, F.; Chen, L.; Xie, K.; Wang, J.; Zhang, G. Transcriptome Analysis of CircRNA and MRNA in Theca Cells during Follicular Development in Chickens. Genes 2020, 11, 489. [Google Scholar] [CrossRef]
- Liu, Y.-D.; Li, Y.; Feng, S.-X.; Ye, D.-S.; Chen, X.; Zhou, X.-Y.; Chen, S.-L. Long Noncoding RNAs: Potential Regulators Involved in the Pathogenesis of Polycystic Ovary Syndrome. Endocrinology 2017, 158, 3890–3899. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Liu, S.; Xing, G.; Wang, F. LncRNA PVT1/MicroRNA-17-5p/PTEN Axis Regulates Secretion of E2 and P4, Proliferation, and Apoptosis of Ovarian Granulosa Cells in PCOS. Mol. Ther. Nucleic Acids 2020, 20, 205–216. [Google Scholar] [CrossRef]
- Li, D.; Wang, X.; Dang, Y.; Zhang, X.; Zhao, S.; Lu, G.; Chan, W.-Y.; Leung, P.C.K.; Qin, Y. LncRNA GCAT1 Is Involved in Premature Ovarian Insufficiency by Regulating P27 Translation in GCs via Competitive Binding to PTBP1. Mol. Ther. Nucleic Acids 2021, 23, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niknafs, Y.S.; Pandian, B.; Iyer, H.K.; Chinnaiyan, A.M.; Iyer, M.K. TACO Produces Robust Multisample Transcriptome Assemblies from RNA-Seq. Nat. Methods 2017, 14, 68–70. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wucher, V.; Legeai, F.; Hédan, B.; Rizk, G.; Lagoutte, L.; Leeb, T.; Jagannathan, V.; Cadieu, E.; David, A.; Lohi, H.; et al. FEELnc: A Tool for Long Non-Coding RNA Annotation and Its Application to the Dog Transcriptome. Nucleic Acids Res. 2017, 45, e57. [Google Scholar] [CrossRef] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Gil, N.; Ulitsky, I. Regulation of Gene Expression by Cis-Acting Long Non-Coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long Noncoding RNAs with Enhancer-like Function in Human Cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Koganti, P.P.; Yao, J. Systematic Identification of Long Intergenic Non-Coding RNAs Expressed in Bovine Oocytes. Reprod. Biol. Endocrinol. 2020, 18, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, J.; Chen, Y.; Li, Z.; Yang, H.; Chen, H.; Yu, Z. Long Non-Coding RNAs in Ovarian Granulosa Cells. J. Ovarian Res. 2020, 13, 63. [Google Scholar] [CrossRef]
- Costermans, N.G.J.; Soede, N.M.; van Tricht, F.; Blokland, M.; Kemp, B.; Keijer, J.; Teerds, K.J. Follicular Fluid Steroid Profile in Sows: Relationship to Follicle Size and Oocyte Quality†. Biol. Reprod. 2020, 102, 740–749. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, G.E.; Bergh, P.A.; Guzman, I.; Masuku, S.; Navot, D. Premature Luteinization Is Not Eliminated by Pituitary Desensitization with Leuprolide Acetate in Women Undergoing Gonadotrophin Stimulation Who Demonstrated Premature Luteinization in a Prior Gonadotrophin-Only Cycle. Hum. Reprod. 1993, 8, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Zheng, Q.; Sui, M.; Zhu, L.; Xu, L.; Zhang, Y.; Liu, Y.; Fang, F.; Chu, M.; Ma, Y.; et al. Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development. Int. J. Mol. Sci. 2019, 20, 3950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, C.; Ma, T.; Hu, S.; Xu, Z.; Zhang, P.; Zhao, X.; Wang, Y.; Yin, H.; Hu, Y.; Fan, X.; et al. Long Non-Coding RNA and MRNA Profile of Liver Tissue During Four Developmental Stages in the Chicken. Front. Genet. 2020, 11, 574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, Z.; Qin, Q.; Nisenblat, V.; Chang, H.-M.; Yu, Y.; Wang, T.; Lu, C.; Yang, M.; Yang, S.; et al. Transcriptome Landscape of Human Folliculogenesis Reveals Oocyte and Granulosa Cell Interactions. Mol. Cell 2018, 72, 1021–1034.e4. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Li, P.; Yang, W.; Ruan, X.; Kiesewetter, K.; Zhu, J.; Cao, H. Integrative Transcriptome Analyses of Metabolic Responses in Mice Define Pivotal LncRNA Metabolic Regulators. Cell Metab. 2016, 24, 627–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donadeu, F.X.; Ginther, O.J. Changes in Concentrations of Follicular Fluid Factors During Follicle Selection in Mares. Biol. Reprod. 2002, 66, 1111–1118. [Google Scholar] [CrossRef] [Green Version]
- Gastal, E.L.; Gastal, M.O.; Ginther, O.J. Experimental Assumption of Dominance by a Smaller Follicle and Associated Hormonal Changes in Mares. Biol. Reprod. 1999, 61, 724–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunzicker-Dunn, M.; Maizels, E.T. FSH Signaling Pathways in Immature Granulosa Cells That Regulate Target Gene Expression: Branching out from Protein Kinase A. Cell Signal. 2006, 18, 1351–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunzicker-Dunn, M.E.; Lopez-Biladeau, B.; Law, N.C.; Fiedler, S.E.; Carr, D.W.; Maizels, E.T. PKA and GAB2 Play Central Roles in the FSH Signaling Pathway to PI3K and AKT in Ovarian Granulosa Cells. Proc. Natl. Acad. Sci. USA 2012, 109, E2979–E2988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.W.; Chung, E.J.; Jung, S.-A.; Lee, J.H. PDGF Stimulation of Müller Cell Proliferation: Contributions of c-JNK and the PI3K/Akt Pathway. Biochem. Biophys. Res. Commun. 2009, 388, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Blumenthal, D.K.; Terry, C.M.; He, Y.; Carlson, M.L.; Cheung, A.K. PDGF-Induced Proliferation in Human Arterial and Venous Smooth Muscle Cells: Molecular Basis for Differential Effects of PDGF Isoforms. J. Cell. Biochem. 2011, 112, 289–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, D.M.; Ko, C.; Jo, M.; Brannstrom, M.; Curry, T.E. Ovulation: Parallels With Inflammatory Processes. Endocr. Rev. 2019, 40, 369–416. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Huang, K.; Hu, S.; Lan, G.; Gan, X.; Gao, S.; Deng, Y.; Hu, J.; Li, L.; Hu, B.; et al. FASN-Mediated Lipid Metabolism Regulates Goose Granulosa Cells Apoptosis and Steroidogenesis. Front. Physiol. 2020, 11, 600. [Google Scholar] [CrossRef]
- Gao, S.; Gan, X.; He, H.; Hu, S.; Deng, Y.; Chen, X.; Li, L.; Hu, J.; Li, L.; Wang, J. Dynamic Characteristics of Lipid Metabolism in Cultured Granulosa Cells from Geese Follicles at Different Developmental Stages. Biosci. Rep. 2019, 39, BSR20192188. [Google Scholar] [CrossRef]
- Plante-Dubé, M.; Picard, C.; Gilbert, I.; Robert, C.; Fievez, V.; Vlaeminck, B.; Belleannée, C.; Gervais, R.; Chouinard, P.Y. Effects of a Dietary Supplement Enriched in Palmitoleic Acid on Fatty Acid Composition of Follicular Fluid, Granulosa Cell Metabolism, and Oocyte Developmental Capacity in Early Lactation Dairy Cows. J. Dairy Sci. 2021, 104, 3693–3706. [Google Scholar] [CrossRef]
- Hu, S.; Gao, S.; Zhu, J.; Gan, X.; Chen, X.; He, H.; Liang, L.; Hu, B.; Hu, J.; Liu, H.; et al. Differential Actions of Diacylglycerol Acyltransferase (DGAT) 1 and 2 in Regulating Lipid Metabolism and Progesterone Secretion of Goose Granulosa Cells. J. Steroid Biochem. Mol. Biol. 2020, 202, 105721. [Google Scholar] [CrossRef]
- Elis, S.; Desmarchais, A.; Maillard, V.; Uzbekova, S.; Monget, P.; Dupont, J. Cell Proliferation and Progesterone Synthesis Depend on Lipid Metabolism in Bovine Granulosa Cells. Theriogenology 2015, 83, 840–853. [Google Scholar] [CrossRef] [PubMed]
- Appasamy, M.; Jauniaux, E.; Serhal, P.; Al-Qahtani, A.; Groome, N.P.; Muttukrishna, S. Evaluation of the Relationship between Follicular Fluid Oxidative Stress, Ovarian Hormones, and Response to Gonadotropin Stimulation. Fertil. Steril. 2008, 89, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Shkolnik, K.; Tadmor, A.; Ben-Dor, S.; Nevo, N.; Galiani, D.; Dekel, N. Reactive Oxygen Species Are Indispensable in Ovulation. Proc. Natl. Acad. Sci. USA 2011, 108, 1462–1467. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Nawani, N.; Malhotra, N.; Malhotra, J.; Patil, M.; Jayakrishnan, K.; Kar, S.; Jirge, P.R.; Mahajan, N. Assisted Reproduction in Polycystic Ovarian Disease: A Multicentric Trial in India. J. Hum. Reprod. Sci. 2013, 6, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Kedem-Dickman, A.; Maman, E.; Yung, Y.; Yerushalmi, G.M.; Hemi, R.; Hanochi, M.; Dor, J.; Hourvitz, A. Anti-Müllerian Hormone Is Highly Expressed and Secreted from Cumulus Granulosa Cells of Stimulated Preovulatory Immature and Atretic Oocytes. Reprod. Biomed. Online 2012, 24, 540–546. [Google Scholar] [CrossRef] [Green Version]
- Direito, A.; Bailly, S.; Mariani, A.; Ecochard, R. Relationships between the Luteinizing Hormone Surge and Other Characteristics of the Menstrual Cycle in Normally Ovulating Women. Fertil. Steril. 2013, 99, 279–285.e3. [Google Scholar] [CrossRef]
- Medzhitov, R. Origin and Physiological Roles of Inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Robker, R.L.; Hennebold, J.D.; Russell, D.L. Coordination of Ovulation and Oocyte Maturation: A Good Egg at the Right Time. Endocrinology 2018, 159, 3209–3218. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Lin, P.-C.; Zhou, S.; Barakat, R.; Bashir, S.T.; Choi, J.M.; Cacioppo, J.A.; Oakley, O.R.; Duffy, D.M.; Lydon, J.P.; et al. Progesterone Receptor Serves the Ovary as a Trigger of Ovulation and a Terminator of Inflammation. Cell Rep. 2020, 31, 107496. [Google Scholar] [CrossRef]
- Sapoznik, S.; Bahar-Shany, K.; Brand, H.; Pinto, Y.; Gabay, O.; Glick-Saar, E.; Dor, C.; Zadok, O.; Barshack, I.; Zundelevich, A.; et al. Activation-Induced Cytidine Deaminase Links Ovulation-Induced Inflammation and Serous Carcinogenesis. Neoplasia 2016, 18, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Lorenz, T.K.; Worthman, C.M.; Vitzthum, V.J. Links among Inflammation, Sexual Activity and Ovulation: Evolutionary Trade-Offs and Clinical Implications. Evol. Med. Public Health 2015, 2015, 304–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shankar, S.P.; Wilson, M.S.; DiVietro, J.A.; Mentink-Kane, M.M.; Xie, Z.; Wynn, T.A.; Druey, K.M. RGS16 Attenuates Pulmonary Th2/Th17 Inflammatory Responses. J. Immunol. 2012, 188, 6347–6356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasson, R.; Rimon, E.; Dantes, A.; Cohen, T.; Shinder, V.; Land-Bracha, A.; Amsterdam, A. Gonadotrophin-induced Gene Regulation in Human Granulosa Cells Obtained from IVF Patients. Modulation of Steroidogenic Genes, Cytoskeletal Genes and Genes Coding for Apoptotic Signalling and Protein Kinases. Mol. Hum. Reprod. 2004, 10, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Derrien, A.; Druey, K.M. RGS16 Function Is Regulated by Epidermal Growth Factor Receptor-Mediated Tyrosine Phosphorylation. J. Biol. Chem. 2001, 276, 48532–48538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, M.; Umehara, T.; Hoshino, Y. Roles of Epidermal Growth Factor (EGF)-like Factor in the Ovulation Process. Reprod. Med. Biol. 2016, 15, 201–216. [Google Scholar] [CrossRef] [PubMed]
- El-Hayek, S.; Demeestere, I.; Clarke, H.J. Follicle-Stimulating Hormone Regulates Expression and Activity of Epidermal Growth Factor Receptor in the Murine Ovarian Follicle. Proc. Natl. Acad. Sci. USA 2014, 111, 16778–16783. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.; Asada, H.; Kizuka, F.; Tamura, I.; Maekawa, R.; Taketani, T.; Sato, S.; Yamagata, Y.; Tamura, H.; Sugino, N. Changes in Histone Modification and DNA Methylation of the StAR and Cyp19a1 Promoter Regions in Granulosa Cells Undergoing Luteinization during Ovulation in Rats. Endocrinology 2013, 154, 458–470. [Google Scholar] [CrossRef]
- Sugino, N. Molecular Mechanisms of Luteinization. Obstet. Gynecol. Sci. 2014, 57, 93–101. [Google Scholar] [CrossRef]
- Wilhelm, L.P.; Wendling, C.; Védie, B.; Kobayashi, T.; Chenard, M.-P.; Tomasetto, C.; Drin, G.; Alpy, F. STARD3 Mediates Endoplasmic Reticulum-to-Endosome Cholesterol Transport at Membrane Contact Sites. EMBO J. 2017, 36, 1412–1433. [Google Scholar] [CrossRef]






| Samples | Raw Reads | Raw Base(G) 1 | Q20(%) 2 | Q30(%) 3 | GC Content(%) 4 |
|---|---|---|---|---|---|
| ZT0_1 | 54,270,570 | 16.28 | 97.56 | 93.34 | 48.82 |
| ZT0_2 | 56,376,040 | 16.91 | 97.34 | 92.95 | 48.92 |
| ZT0_3 | 60,354,655 | 18.11 | 97.13 | 92.45 | 47.84 |
| ZT12_1 | 67,301,276 | 20.19 | 97.35 | 92.91 | 51.37 |
| ZT12_2 | 47,927,165 | 14.38 | 97.44 | 93.17 | 52.69 |
| ZT12_3 | 61,112,368 | 18.33 | 97.44 | 93.17 | 55.09 |
| ZT20_1 | 58,567,244 | 17.57 | 97.59 | 93.41 | 48.12 |
| ZT20_2 | 53,311,100 | 15.99 | 97.53 | 93.27 | 47.93 |
| ZT20_3 | 52,926,730 | 15.88 | 97.17 | 92.61 | 54.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Deng, X.; Hu, S.; Cui, Z.; Ning, Z.; Gui, T.; Zhao, X.; Li, D.; Wang, Y.; Yin, H.; et al. Systematic Analysis of Long Noncoding RNA and mRNA in Granulosa Cells during the Hen Ovulatory Cycle. Animals 2021, 11, 1533. https://doi.org/10.3390/ani11061533
Li L, Deng X, Hu S, Cui Z, Ning Z, Gui T, Zhao X, Li D, Wang Y, Yin H, et al. Systematic Analysis of Long Noncoding RNA and mRNA in Granulosa Cells during the Hen Ovulatory Cycle. Animals. 2021; 11(6):1533. https://doi.org/10.3390/ani11061533
Chicago/Turabian StyleLi, Liang, Xun Deng, Silu Hu, Zhifu Cui, Zifan Ning, Taotao Gui, Xiaoling Zhao, Diyan Li, Yan Wang, Huadong Yin, and et al. 2021. "Systematic Analysis of Long Noncoding RNA and mRNA in Granulosa Cells during the Hen Ovulatory Cycle" Animals 11, no. 6: 1533. https://doi.org/10.3390/ani11061533
APA StyleLi, L., Deng, X., Hu, S., Cui, Z., Ning, Z., Gui, T., Zhao, X., Li, D., Wang, Y., Yin, H., Ye, L., Tian, Y., Zhang, Y., Li, H., & Zhu, Q. (2021). Systematic Analysis of Long Noncoding RNA and mRNA in Granulosa Cells during the Hen Ovulatory Cycle. Animals, 11(6), 1533. https://doi.org/10.3390/ani11061533






