Comparative Metagenomic Analysis of Chicken Gut Microbial Community, Function, and Resistome to Evaluate Noninvasive and Cecal Sampling Resources
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chickens, Diets, and Sampling
2.2. DNA Extraction and Library Preparation and Sequencing
2.3. Metagenome Assembly, Gene Prediction, and Abundance Analysis
2.4. Taxonomy Prediction and Gene Function Analysis
2.5. Statistical Analysis
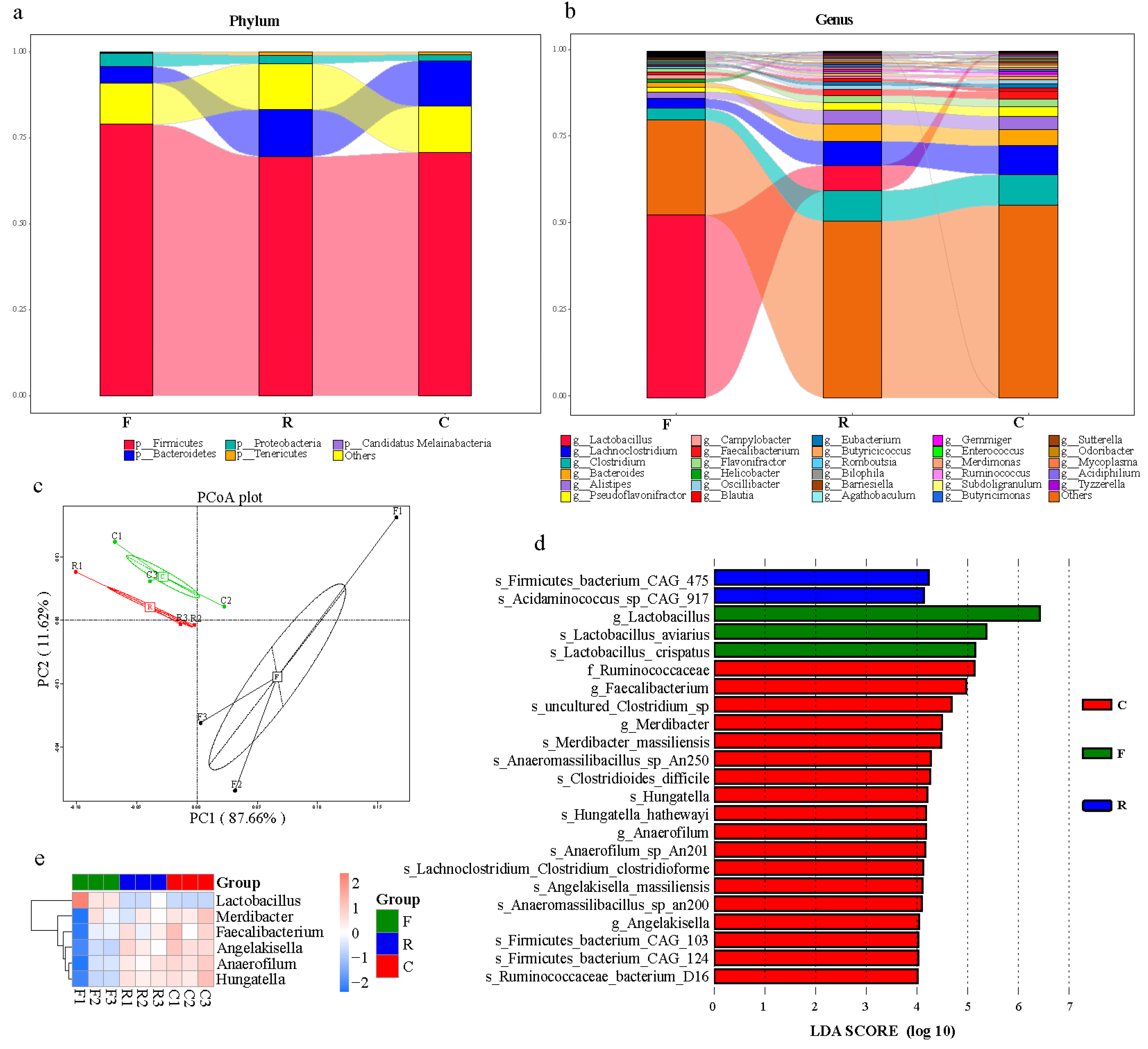
3. Results
3.1. Sequencing, Assembly, and Microbial Taxonomy
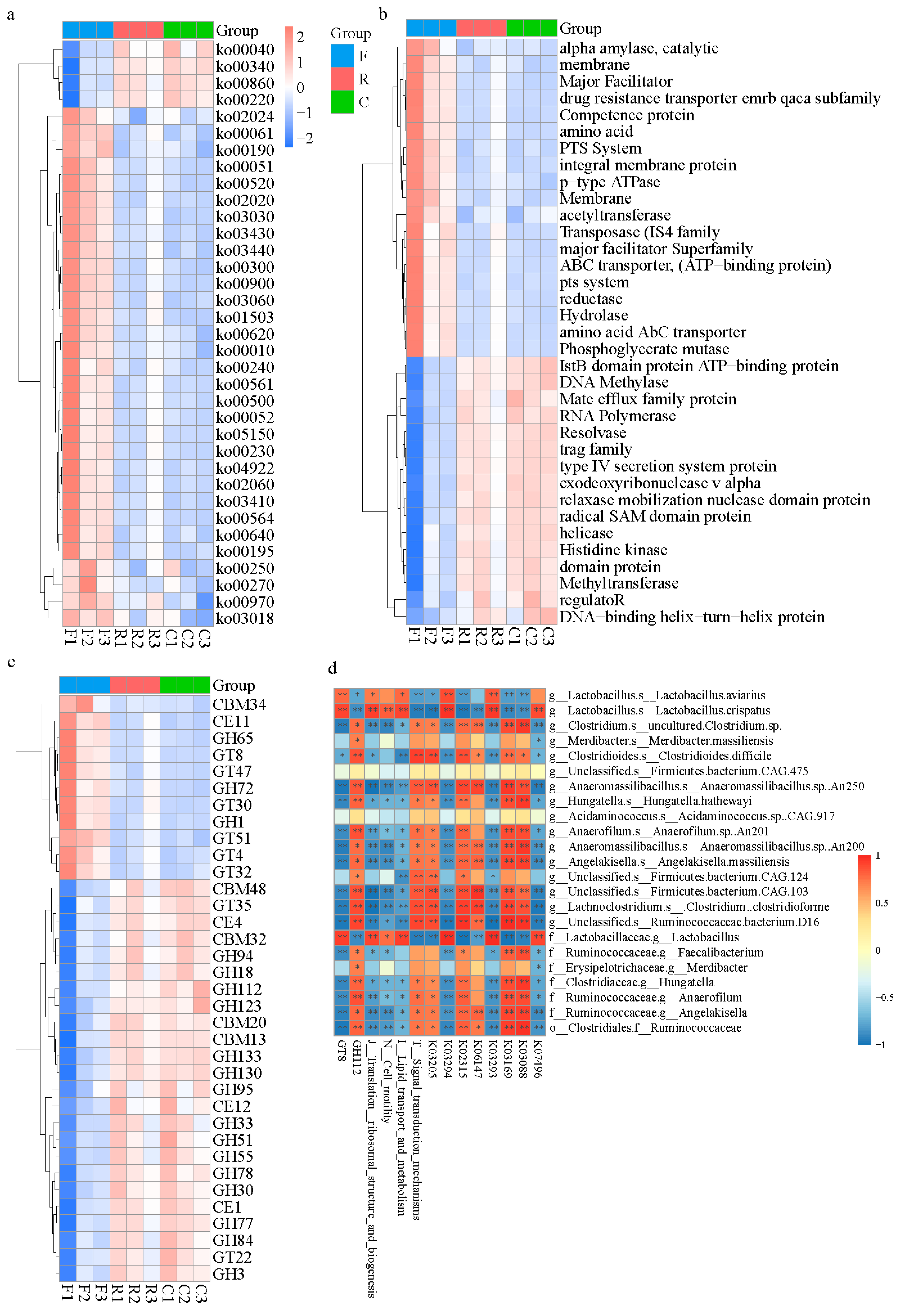
3.2. Bacterial Functional Analysis
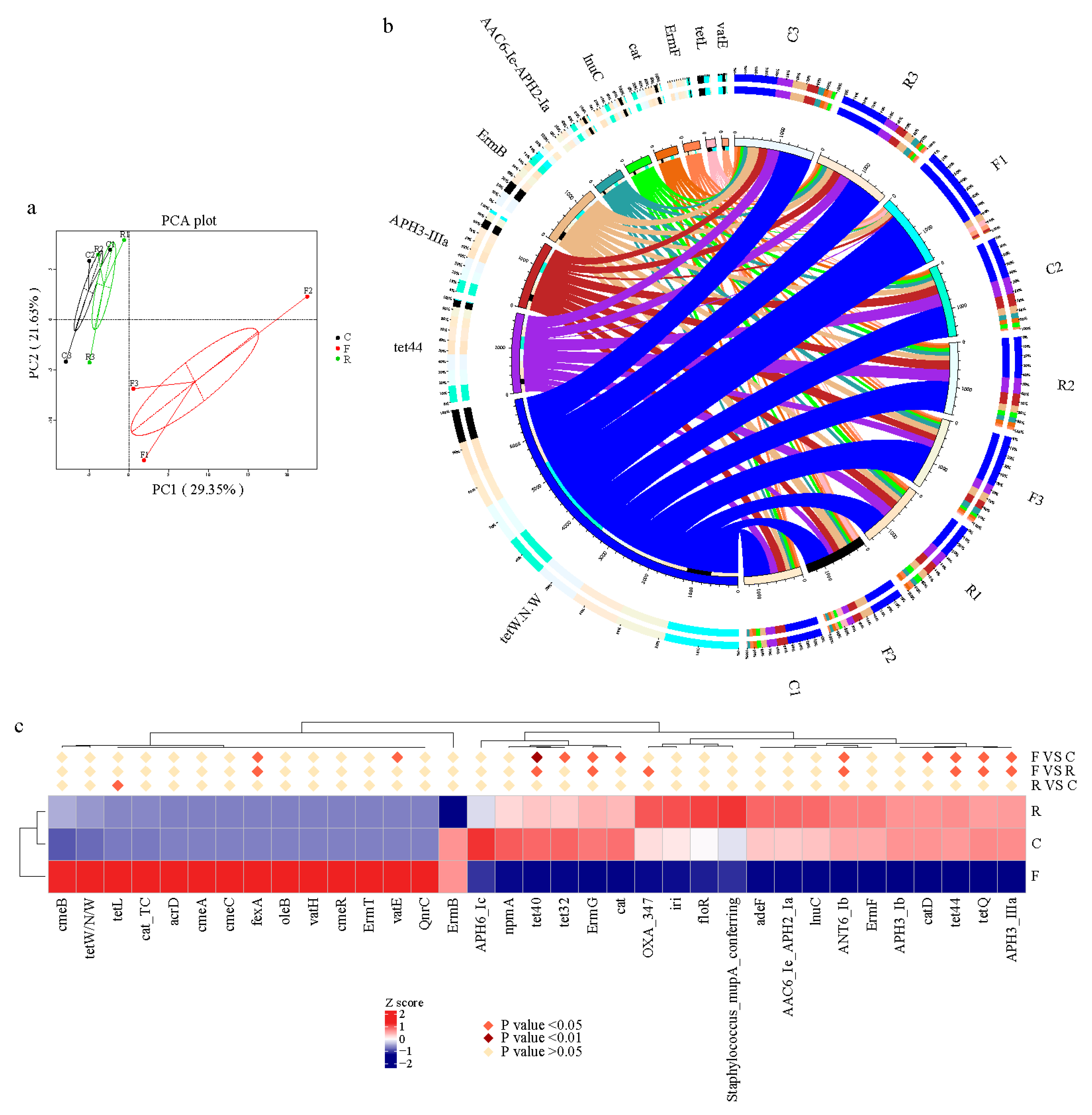
3.3. Diversity and Abundance of Antibiotic Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, C.; Liu, H.; Zhang, Y.; Lu, C. Comparative analysis of the gut microbiota of hornbill and toucan in captivity. Microbiologyopen 2018, 8, e00786. [Google Scholar] [CrossRef] [PubMed]
- Lyte, M. The microbial organ in the gut as a driver of homeostasis and disease. Med. Hypotheses 2010, 74, 634–638. [Google Scholar] [CrossRef] [PubMed]
- Hartstra, A.V.; Bouter, K.E.; Bäckhed, F.; Nieuwdorp, M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care 2015, 38, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Perruzza, L.; Strati, F.; Gargari, G.; D’Erchia, A.M.; Fosso, B.; Pesole, G.; Guglielmetti, S.; Grassi, F. Enrichment of intestinal Lactobacillus by enhanced secretory IgA coating alters glucose homeostasis in P2rx7(-/-) mice. Sci. Rep. 2019, 9, 9315. [Google Scholar] [CrossRef]
- Lagier, J.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.; Fournier, P.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef]
- Tan, Z.; Luo, L.; Wang, X.; Wen, Q.; Zhou, L.; Wu, K. Characterization of the cecal microbiome composition of Wenchang chickens before and after fattening. PLoS ONE 2019, 14, e225692. [Google Scholar] [CrossRef]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207. [Google Scholar]
- Reyman, M.; van Houten, M.A.; Arp, K.; Sanders, E.A.M.; Bogaert, D. Rectal swabs are a reliable proxy for faecal samples in infant gut microbiota research based on 16S-rRNA sequencing. Sci. Rep. 2019, 9, 16072. [Google Scholar] [CrossRef]
- Stanley, D.; Geier, M.S.; Chen, H.; Hughes, R.J.; Moore, R.J. Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences. BMC Microbiol. 2015, 15, 51. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, A.; Iimuro, M.; Sato, R.; Yamazaki, J.; Inomata, T. Effect of midazolam and butorphanol premedication on inhalant isoflurane anesthesia in mice. Exp. Anim. 2015, 64, 14–73. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Duan, X.; Peng, Y.; Rui, Y. Molecular characteristics of carbapenem-resistant Acinetobacter spp. from clinical infection samples and fecal survey samples in Southern China. BMC Infect. Dis. 2019, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Borhani, K.; Ahmadi, A.; Rahimi, F.; Pourshafie, M.R.; Talebi, M. Determination of Vancomycin Resistant Enterococcus faecium Diversity in Tehran Sewage Using Plasmid Profile, Biochemical Fingerprinting and Antibiotic Resistance. Jundishapur J. Microbiol. 2014, 7, e8951. [Google Scholar] [CrossRef] [PubMed]
- Kozik, A.J.; Nakatsu, C.H.; Chun, H.; Jones-Hall, Y.L. Comparison of the fecal, cecal, and mucus microbiome in male and female mice after TNBS-induced colitis. PLoS ONE 2019, 14, e225079. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Lennon, G.; O’Sullivan, O.; Docherty, N.; Balfe, A.; Maguire, A.; Mulcahy, H.E.; Doherty, G.; O’Donoghue, D.; Hyland, J.; et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015, 64, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Sun, C.; Zheng, J.; Wen, C.; Ji, C.; Zhang, D.; Chen, Y.; Hou, Z.; Yang, N. Efficacy of Fecal Sampling as a Gut Proxy in the Study of Chicken Gut Microbiota. Front. Microbiol. 2019, 10, 2126. [Google Scholar] [CrossRef]
- Yasuda, K.; Oh, K.; Ren, B.; Tickle, T.L.; Franzosa, E.A.; Wachtman, L.M.; Miller, A.D.; Westmoreland, S.V.; Mansfield, K.G.; Vallender, E.J.; et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 2015, 17, 385–391. [Google Scholar] [CrossRef]
- Wu, H.; Xing, Y.; Sun, H.; Mao, X. Gut microbial diversity in two insectivorous bats: Insights into the effect of different sampling sources. Microbiologyopen 2019, 8, e00670. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, Y.; Liu, S.; Huang, J.; Zhai, Z.; He, C.; Ding, J.; Wang, J.; Wang, H.; Fan, W.; et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS ONE 2015, 10, e0117441. [Google Scholar]
- Panasevich, M.R.; Wankhade, U.D.; Chintapalli, S.V.; Shankar, K.; Rector, R.S. Cecal versus fecal microbiota in Ossabaw swine and implications for obesity. Physiol. Genom. 2018, 50, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Zhang, Y.; Xiao, K.; Jiang, F.; Wang, H.; Tang, D.; Liu, D.; Liu, B.; Liu, Y.; He, X.; et al. The chicken gut metagenome and the modulatory effects of plant-derived benzylisoquinoline alkaloids. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Rychlik, I. Composition and Function of Chicken Gut Microbiota. Animals 2020, 10, 103. [Google Scholar] [CrossRef]
- Liu, G.; Bou, G.; Su, S.; Xing, J.; Qu, H.; Zhang, X.; Wang, X.; Zhao, Y.; Dugarjaviin, M. Microbial diversity within the digestive tract contents of Dezhou donkeys. PLoS ONE 2019, 14, e0226186. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 1985, 5, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Zhang, J.; Sethuraman, V.; Cui, G.; Yi, X.; Zhong, G. Transcriptome analysis of Spodoptera frugiperda Sf9 cells reveals putative apoptosis-related genes and a preliminary apoptosis mechanism induced by azadirachtin. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Oakley, B.B.; Kogut, M.H. Spatial and temporal changes in the broiler chicken cecal and fecal microbiomes and correlations of bacterial taxa with cytokine gene expression. Front. Vet. Sci. 2016, 3, 11. [Google Scholar] [CrossRef]
- Whitfield-Cargile, C.M.; Cohen, N.D.; He, K.; Ivanov, I.; Goldsby, J.S.; Chamoun-Emanuelli, A.; Weeks, B.R.; Davidson, L.A.; Chapkin, R.S. The non-invasive exfoliated transcriptome (exfoliome) reflects the tissue-level transcriptome in a mouse model of NSAID enteropathy. Sci. Rep. 2017, 7, 1–13. [Google Scholar]
- Wen, C.; Yan, W.; Sun, C.; Ji, C.; Zhou, Q.; Zhang, D.; Zheng, J.; Yang, N. The gut microbiota is largely independent of host genetics in regulating fat deposition in chickens. ISME J. 2019, 13, 1422–1436. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, L.; Shao, D.; Wang, Q.; Wu, Y.; Han, Y.; Shi, S. Selectived and Reshaped Early Dominant Microbial Community in the Cecum with Similar Proportions and Better Homogenization and Species Diversity Due to Organic Acids as AGP Alternatives Mediate Their Effects on Broilers Growth. Front. Microbiol. 2020, 10, 2948. [Google Scholar] [CrossRef]
- Yausheva, E.; Miroshnikov, S.; Sizova, E. Intestinal microbiome of broiler chickens after use of nanoparticles and metal salts. Environ. Sci. Pollut. Res. 2018, 25, 18109–18120. [Google Scholar] [CrossRef]
- Borda-Molina, D.; Seifert, J.; Camarinha-Silva, A. Current perspectives of the chicken gastrointestinal tract and its microbiome. Comput. Struct. Biotechnol. J. 2018, 16, 131–139. [Google Scholar] [CrossRef]
- Videnska, P.; Rahman, M.M.; Faldynova, M.; Babak, V.; Matulova, M.E.; Prukner-Radovcic, E.; Krizek, I.; Smole-Mozina, S.; Kovac, J.; Szmolka, A.; et al. Characterization of egg laying hen and broiler fecal microbiota in poultry farms in Croatia, Czech Republic, Hungary and Slovenia. PLoS ONE 2014, 9, e110076. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Estellé, J.; Kiilerich, P.; Ramayo-Caldas, Y.; Xia, Z.; Feng, Q.; Liang, S.; Pedersen, A.Ø.; Kjeldsen, N.J.; Liu, C. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016, 1, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, S.; Ulloa-Martínez, M.; Martínez-Rojano, H.; Galván-Rodríguez, F.M.; Miranda-Brito, C.; Romano, M.C.; Piña-Escobedo, A.; Pizano-Zárate, M.L.; Hoyo-Vadillo, C.; García-Mena, J. Study of the diversity and short-chain fatty acids production by the bacterial community in overweight and obese Mexican children. Eur. J. Clin. Microbiol. 2015, 34, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Graeme, A.O.; Reynolds, N.; Smith, A.R.; Kennedy, A.; Macfarlane, G.T. Effect of pH and antibiotics on microbial overgrowth in the stomachs and duodena of patients undergoing percutaneous endoscopic gastrostomy feeding. J. Clin. Microbiol. 2005, 43, 3059–3065. [Google Scholar]
- Espey, M.G. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic. Biol. Med. 2013, 55, 130–140. [Google Scholar] [CrossRef]
- Sekelja, M.; Rud, I.; Knutsen, S.H.; Denstadli, V.; Westereng, B.; Naes, T.; Rudi, K. Abrupt temporal fluctuations in the chicken fecal microbiota are explained by its gastrointestinal origin. Appl. Environ. Microbiol. 2012, 78, 2941–2948. [Google Scholar] [CrossRef]
- Sun, H.; Tang, J.W.; Fang, C.L.; Yao, X.H.; Wu, Y.F.; Wang, X.; Feng, J. Molecular analysis of intestinal bacterial microbiota of broiler chickens fed diets containing fermented cottonseed meal. Poult. Sci. 2013, 92, 392–401. [Google Scholar] [CrossRef]
- Morris, L.; Thompson, R.B.; Heller, V.G. Crude fiber in chicken rations. Poult. Sci. 1932, 11, 219–225. [Google Scholar] [CrossRef]
- Zechiedrich, E.L.; Khodursky, A.B.; Cozzarelli, N.R. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination inEscherichia coli. Gene Dev. 1997, 11, 2580–2592. [Google Scholar] [CrossRef]
- Lopez, C.R.; Yang, S.; Deibler, R.W.; Ray, S.A.; Pennington, J.M.; DiGate, R.J.; Hastings, P.J.; Rosenberg, S.M.; Zechiedrich, E.L. A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol. Microbiol. 2005, 58, 80–101. [Google Scholar] [CrossRef] [PubMed]
- Backert, S.; Meyer, T.F. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 2006, 9, 207–217. [Google Scholar] [CrossRef]
- Schmid, M.C.; Schulein, R.; Dehio, M.; Denecker, G.; Carena, I.; Dehio, C. The VirB type IV secretion system of Bartonella henselae mediates invasion, proinflammatory activation and antiapoptotic protection of endothelial cells. Mol. Microbiol. 2004, 52, 81–92. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, G.; Siegel, P.; He, C.; Wang, H.; Zhao, W.; Zhai, Z.; Tian, F.; Zhao, J.; Zhang, H.; et al. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci. Rep. 2013, 3, 1–6. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Liu, H.; Brown, M.A.; Qiao, S. Dietary protein and gut microbiota composition and function. Curr. Protein Pept. Sci. 2019, 20, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, S.P.; Stokes, H.W.; Chowdhury, P.R. Mobile elements, zoonotic pathogens and commensal bacteria: Conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front. Microbiol. 2013, 4, 86. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M. Prevention of Atherosclerosis by the Induction of Microbial Polyamine Production in the Intestinal Lumen. Biol. Pharm. Bull. 2020, 43, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Chen, H.; Hahn, M.G.; Mohnen, D.; Xu, Y. Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant Physiol. 2010, 153, 1729–1746. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, G.; Zhang, R.; Shen, Y.; Tyrrell, J.M.; Huang, X.; Zhou, H.; Lei, L.; Li, H.; Doi, Y.; et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: An epidemiological and clinical study. Lancet Infect. Dis. 2017, 17, 390–399. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, R.; Li, J.; Wu, Z.; Yin, W.; Schwarz, S.; Tyrrell, J.M.; Zheng, Y.; Wang, S.; Shen, Z.; et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Wang, Y.; Sun, Y.; Ma, L.; Zeng, Q.; Jiang, X.; Li, A.; Zeng, Z.; Zhang, T. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome 2018, 6, 1–11. [Google Scholar] [CrossRef]
- Lu, X.; Tang, B.; Zhang, Q.; Liu, L.; Fan, R.; Zhang, Z. The presence of Cu facilitates adsorption of tetracycline (TC) onto water hyacinth roots. Int. J. Environ. Res. Public Health 2018, 15, 1982. [Google Scholar] [CrossRef]
- Woegerbauer, M.; Zeinzinger, J.; Springer, B.; Hufnagl, P.; Indra, A.; Korschineck, I.; Hofrichter, J.; Kopacka, I.; Fuchs, R.; Steinwider, J.; et al. Prevalence of the aminoglycoside phosphotransferase genes aph (3′)-IIIa and aph (3′)-IIa in Escherichia coli, Enterococcus faecalis, Enterococcus faecium, Pseudomonas aeruginosa, Salmonella enterica subsp. enterica and Staphylococcus aureus isolates in Austria. J. Med. Microbiol. 2014, 63, 210–217. [Google Scholar] [PubMed]
- Kehrenberg, C.; Schwarz, S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006, 50, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, K.; Hu, Y.; Wu, S.; Shi, S. Comparative Metagenomic Analysis of Chicken Gut Microbial Community, Function, and Resistome to Evaluate Noninvasive and Cecal Sampling Resources. Animals 2021, 11, 1718. https://doi.org/10.3390/ani11061718
Kang K, Hu Y, Wu S, Shi S. Comparative Metagenomic Analysis of Chicken Gut Microbial Community, Function, and Resistome to Evaluate Noninvasive and Cecal Sampling Resources. Animals. 2021; 11(6):1718. https://doi.org/10.3390/ani11061718
Chicago/Turabian StyleKang, Kelang, Yan Hu, Shu Wu, and Shourong Shi. 2021. "Comparative Metagenomic Analysis of Chicken Gut Microbial Community, Function, and Resistome to Evaluate Noninvasive and Cecal Sampling Resources" Animals 11, no. 6: 1718. https://doi.org/10.3390/ani11061718
APA StyleKang, K., Hu, Y., Wu, S., & Shi, S. (2021). Comparative Metagenomic Analysis of Chicken Gut Microbial Community, Function, and Resistome to Evaluate Noninvasive and Cecal Sampling Resources. Animals, 11(6), 1718. https://doi.org/10.3390/ani11061718






