Emergence and Spread of Cephalosporinases in Wildlife: A Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
Search Strategy
3. Results and Discussion
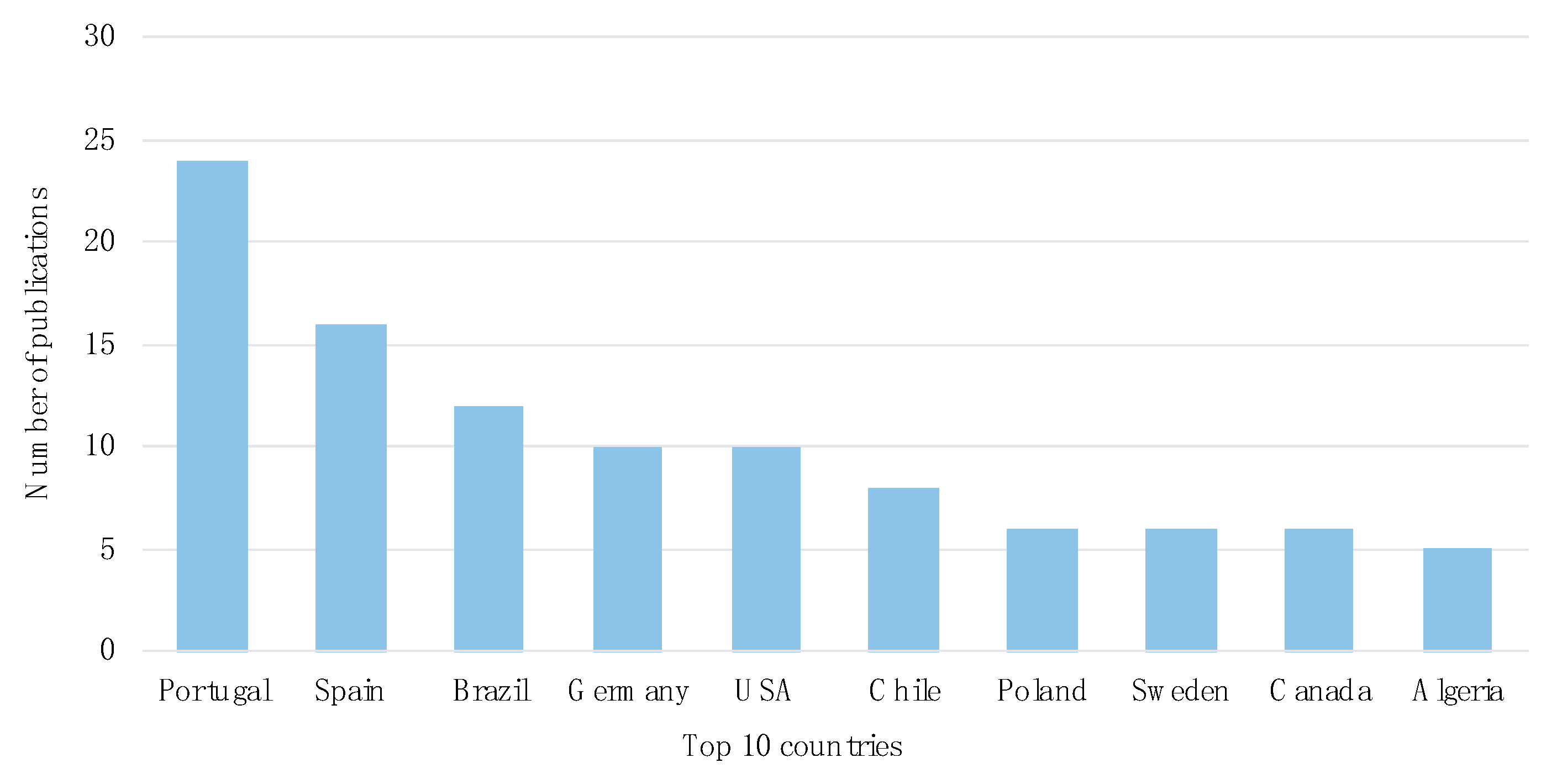
3.1. Temporal and Geographical Research Trends
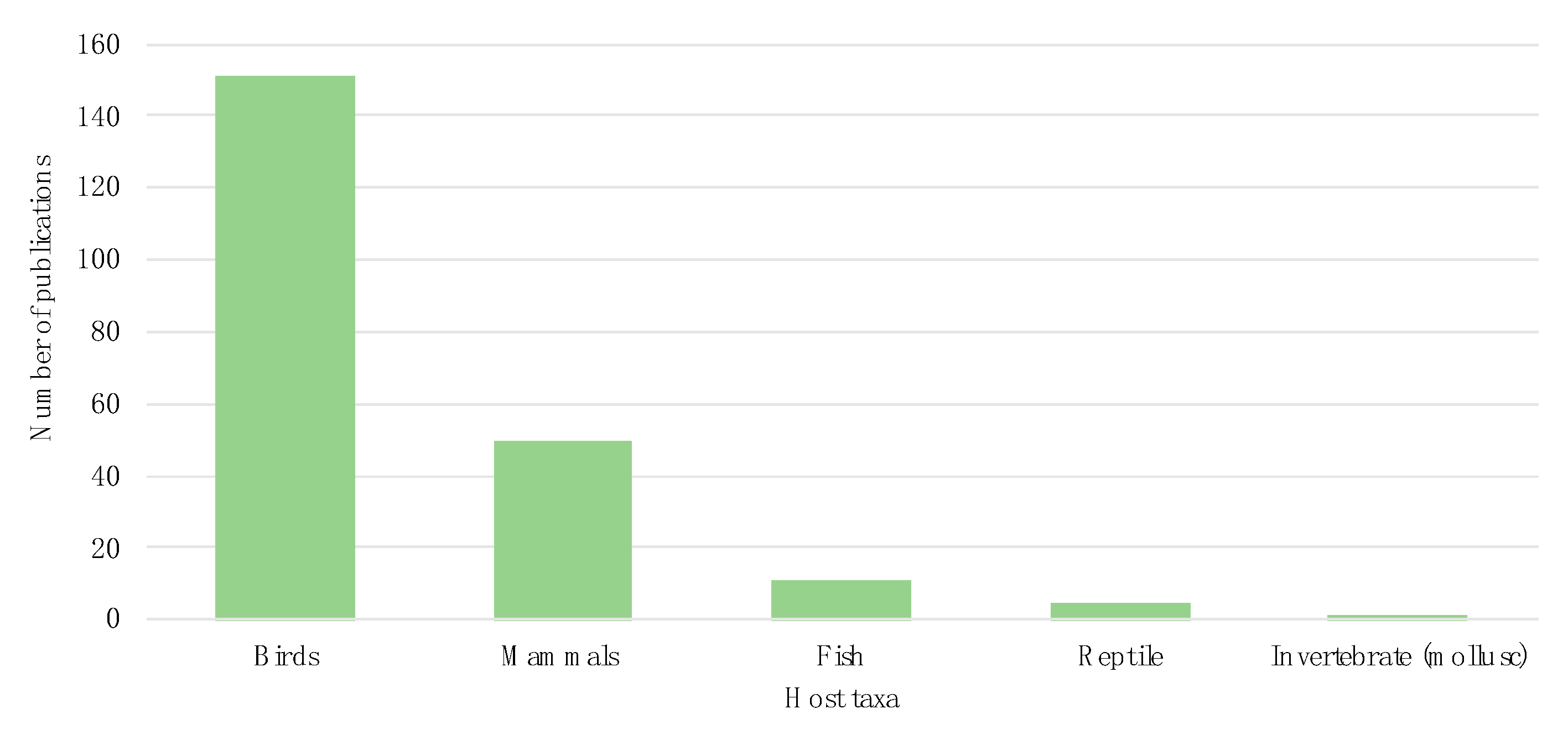
3.2. Host Taxonomy
3.3. Cephalosporinase Diversity in Wildlife Hosts
3.4. Wildlife and Cephalosporinase-Integrative Analyses
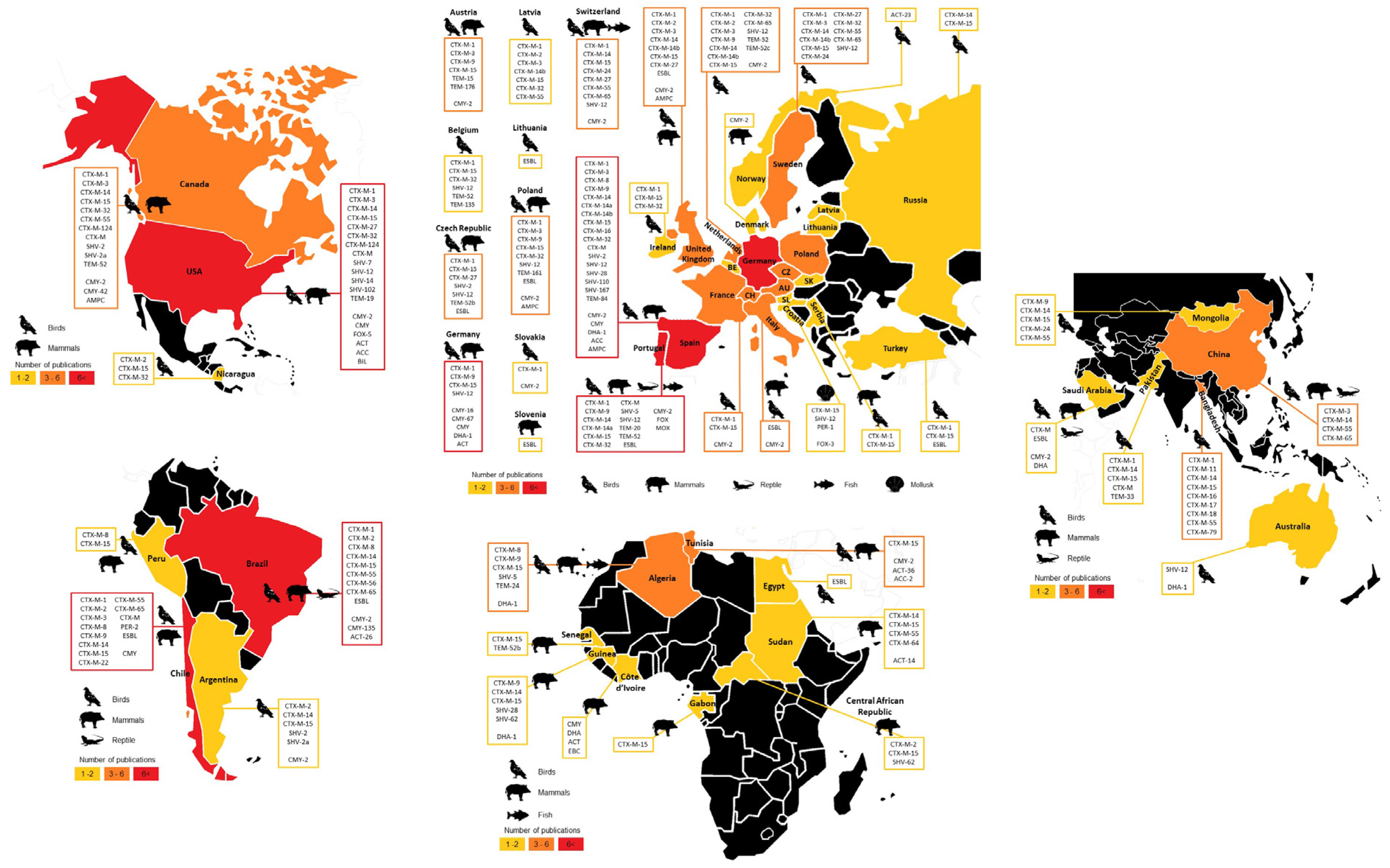
3.4.1. Geographical Overview
3.4.2. Most Disseminated Cephalosporinases
3.4.3. Cephalosporinases and Clonality
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Nordmann, P.; Poirel, L.; Dortet, L. Global Spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Livermore, D.M.; Warner, M.; Hall, L.M.C.; Enne, V.I.; Projan, S.J.; Dunman, P.M.; Wooster, S.L.; Harrison, G. Antibiotic resistance in bacteria from magpies (Pica pica) and rabbits (Oryctolagus cuniculus) from west Wales. Environ. Microbiol. 2001, 3, 658–661. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance—The need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- De Been, M.; Lanza, V.F.; de Toro, M.; Scharringa, J.; Dohmen, W.; Du, Y.; Hu, J.; Lei, Y.; Li, N.; Tooming-Klunderud, A.; et al. Dissemination of Cephalosporin Resistance Genes between Escherichia coli Strains from Farm Animals and Humans by Specific Plasmid Lineages. PLoS Genet. 2014, 10, e1004776. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Dorado-García, A.; van Duijkeren, E.; van den Bunt, G.; Dierikx, C.M.; Bonten, M.J.M.; Bootsma, M.C.J.; Schmitt, H.; Hald, T.; Evers, E.G.; et al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: A population-based modelling study. Lancet Planet. Health 2019, 3, e357–e369. [Google Scholar] [CrossRef]
- Meini, S.; Tascini, C.; Cei, M.; Sozio, E.; Rossolini, G.M. AmpC β-lactamase-producing Enterobacterales: What a clinician should know. Infection 2019, 47, 363–375. [Google Scholar] [CrossRef]
- World Health Organization. WHO List of Critically Important Antimicrobials for Human Medicine (WHO CIA List) 6th Rev [Internet]. World Health Organization. 2017. Available online: http://who.int/foodsafety/publications/antimicrobials-fifth/en/ (accessed on 11 August 2020).
- Berg, E.S.; Wester, A.L.; Ahrenfeldt, J.; Mo, S.S.; Slettemeås, J.S.; Steinbakk, M.; Samuelsen, Ø.; Grude, N.; Simonsen, G.S.; Løhr, I.H.; et al. Norwegian patients and retail chicken meat share cephalosporin-resistant Escherichia coli and IncK/blaCMY-2 resistance plasmids. Clin. Microbiol. Infect. 2017, 23, 407.e9–407.e15. [Google Scholar] [CrossRef] [PubMed]
- Chambers, L.; Yang, Y.; Littier, H.; Ray, P.; Zhang, T.; Pruden, A.; Strickland, M.; Knowlton, K. Metagenomic Analysis of Antibiotic Resistance Genes in Dairy Cow Feces following Therapeutic Administration of Third Generation Cephalosporin. PLoS ONE 2015, 10, e0133764. [Google Scholar] [CrossRef] [PubMed]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial Food Animal Production, Antimicrobial Resistance, and Human Health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Tausova, D.; Dolejska, M.; Cizek, A.; Hanusova, L.; Hrusakova, J.; Svoboda, O.; Camlik, G.; Literak, I. Escherichia coli with extended-spectrum β-lactamase and plasmid-mediated quinolone resistance genes in great cormorants and mallards in Central Europe. J. Antimicrob. Chemother. 2012, 67, 1103–1107. [Google Scholar] [CrossRef]
- Torres, R.T.; Carvalho, J.; Cunha, M.V.; Serrano, E.; Palmeira, J.D.; Fonseca, C. Temporal and geographical research trends of antimicrobial resistance in wildlife—A bibliometric analysis. ONE Health 2020, 11, 100198. [Google Scholar] [CrossRef] [PubMed]
- Liakopoulos, A.; Olsen, B.; Geurts, Y.; Artursson, K.; Berg, C.; Mevius, D.J.; Bonnedahl, J. Molecular characterization of extended-spectrum-cephalosporin-resistant Enterobacteriaceae from wild kelp gulls in South America. Antimicrob. Agents Chemother. 2016, 60, 6924–6927. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Hasman, H.; Olsen, I.; Sørensen, G. International Spread of blaCMY-2-Mediated Cephalosporin Resistance in a Multiresistant Salmonella enterica Serovar Heidelberg Isolate Stemming from the Importation of a Boar by Denmark from Canada. Antimicrob. Agents Chemother. 2004, 48, 1916–1917. [Google Scholar] [CrossRef] [PubMed]
- Arezzo, A.C.; Salehi, H.; Yunus, M.M.; Farhadi, H.; Fooladi, M.; Farhadi, M.; Ale Ebrahim, N. A comparison between two main academic literature collections: Web of science and scopus databases. Asian Soc. Sci. 2013, 9, 18–26. [Google Scholar]
- George, A. Antimicrobial resistance, trade, food safety and security. One Health 2018, 5, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Gilliver, M.A.; Bennett, M.; Begon, M.; Hazel, S.M.; Hart, C.A. Antibiotic resistance found in wild rodents. Nature 1999, 401, 233–234. [Google Scholar] [CrossRef]
- Österblad, M.; Norrdahl, K.; Korpimäki, E.; Huovinen, P. How wild are wild mammals? Nature 2001, 409, 37–38. [Google Scholar] [CrossRef]
- Ahlstrom, C.A.; van Toor, M.L.; Woksepp, H.; Chandler, J.C.; Reed, J.A.; Reeves, A.B.; Waldenström, J.; Franklin, A.B.; Douglas, D.C.; Bonnedahl, J.; et al. Evidence for continental-scale dispersal of antimicrobial resistant bacteria by landfill-foraging gulls. Sci. Total Environ. 2021, 764, 144551. [Google Scholar] [CrossRef]
- Skarżyńska, M.; Zaja̧c, M.; Bomba, A.; Bocian, Ł.; Kozdruń, W.; Polak, M.; Wia̧cek, J.; Wasyl, D. Antimicrobial Resistance Glides in the Sky—Free-Living Birds as a Reservoir of Resistant Escherichia coli with Zoonotic Potential. Front. Microbiol. 2021, 12, 673. [Google Scholar] [CrossRef]
- Vittecoq, M.; Godreuil, S.; Prugnolle, F.; Durand, P.; Brazier, L.; Renaud, N.; Arnal, A.; Aberkane, S.; Jean-Pierre, H.; Gauthier-Clerc, M.; et al. Antimicrobial resistance in wildlife. J. Appl. Ecol. 2016, 53, 519–529. [Google Scholar] [CrossRef]
- Dolejska, M.; Literak, I. Wildlife Is Overlooked in the Epidemiology of Medically Important Antibiotic-Resistant Bacteria. Antimicrob. Agents Chemother. 2019, 63, e01167-19. [Google Scholar] [CrossRef]
- Franklin, A.B.; Ramey, A.M.; Bentler, K.T.; Barrett, N.L.; McCurdy, L.M.; Ahlstrom, C.A.; Bonnedahl, J.; Shriner, S.A.; Chandler, J.C. Gulls as Sources of Environmental Contamination by Colistin-resistant Bacteria. Sci. Rep. 2020, 10, 4408. [Google Scholar] [CrossRef] [PubMed]
- UNECE; FAO. Game Meat-Production and Trade in the UNECE Region; FAO: Roma, Italy, 2018; pp. 1–55. [Google Scholar]
- Coolen, J.P.M.; Den Drijver, E.P.M.; Kluytmans, J.A.J.W.; Verweij, J.J.; Lamberts, B.A.; Soer, J.A.C.J.; Verhulst, C.; Wertheim, H.F.L.; Kolwijck, E. Development of an algorithm to discriminate between plasmid- and chromosomal-mediated AmpC β-lactamase production in Escherichia coli by elaborate phenotypic and genotypic characterization. J. Antimicrob. Chemother. 2019, 74, 3481–3488. [Google Scholar] [CrossRef]
- Dolejska, M.; Papagiannitsis, C.C. Plasmid-mediated resistance is going wild. Plasmid 2018, 99, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.; Laupland, K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Palmeira, J.D.; Ferreira, H. Extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in cattle production—A threat around the world. Heliyon 2020, 6, e03206. [Google Scholar] [CrossRef]
- Agnew, A.; Wang, J.; Fanning, S.; Bearhop, S.; McMahon, B.J. Insights into antimicrobial resistance among long distance migratory East Canadian High Arctic light-bellied Brent geese (Branta bernicla hrota). Ir. Vet. J. 2016, 69, 13. [Google Scholar] [CrossRef] [PubMed]
- Palmeira, J.D.; Ferreira, H.; Madec, J.Y.; Haenni, M. Draft genome of a ST443 mcr-1- and blaCTX-M-2-carrying Escherichia coli from cattle in Brazil. J. Glob. Antimicrob. Resist. 2018, 13, 269–270. [Google Scholar] [CrossRef] [PubMed]
- Palmeira, J.D.; Haenni, M.; Metayer, V.; Madec, J.-Y.; Ferreira, H. Epidemic spread of IncI1/pST113 plasmid carrying the Extended-Spectrum β-Lactamase (ESBL) blaCTX-M-8 gene in Escherichia coli of Brazilian cattle. Vet. Microbiol. 2020, 243, 108629. [Google Scholar] [CrossRef]
- Furlan, J.P.R.; Savazzi, E.A.; Stehling, E.G. Widespread high-risk clones of multidrug-resistant extended-spectrum β-lactamase-producing Escherichia coli B2-ST131 and F-ST648 in public aquatic environments. Int. J. Antimicrob. Agents 2020, 56, 106040. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jiang, Z.-G.; Xia, L.-N.; Shen, J.-Z.; Dai, L.; Wang, Y.; Huang, S.-Y.; Wu, G.-M. Characterization of antimicrobial resistance and molecular determinants of β-lactamase in Escherichia coli isolated from chickens in China during 1970–2007. Vet. Microbiol. 2010, 144, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Altayb, H.N.; Siddig, M.A.M.; El Amin, N.M.; Hashim, A.I.; Mukhtar, M.M. Molecular Characterization of CTX-M ESBLs among Pathogenic Enterobacteriaceae isolated from different regions in Sudan. Glob. Adv. Res. J. Microbiol. 2018, 7, 040–046. [Google Scholar]
- Schaumburg, F.; Alabi, A.; Kokou, C.; Grobusch, M.P.; Köck, R.; Kaba, H.; Becker, K.; Adegnika, A.A.; Kremsner, P.G.; Peters, G.; et al. High burden of extended-spectrum β-lactamase-producing Enterobacteriaceae in Gabon. J. Antimicrob. Chemother. 2013, 68, 2140–2143. [Google Scholar] [CrossRef]
- Frank, T.; Arlet, G.; Gautier, V.; Talarmin, A.; Bercion, R. Extended-spectrum β-lactamase-producing Enterobacteriaceae, Central African Republic. Emerg. Infect. Dis. 2006, 12, 863–865. [Google Scholar] [CrossRef]
- Zhou, J.; Li, C.; Liu, X.; Chiu, M.C.; Zhao, X.; Wang, D.; Wei, Y.; Lee, A.; Zhang, A.J.; Chu, H.; et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020, 26, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Shamsuzzaman, S. Emergence of carbapenemase-producing urinary isolates at a tertiary care hospital in Dhaka, Bangladesh. Tzu Chi Med. J. 2016, 28, 94–98. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J.D. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: The worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 2010, 35, 316–321. [Google Scholar] [CrossRef]
- Sheng, W.H.; Badal, R.E.; Hsueh, P.R. Distribution of extended-spectrum β-lactamases, AmpC β-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: Results of the study for monitoring antimicrobial resistance trends (SMART). Antimicrob. Agents Chemother. 2013, 57, 2981–2988. [Google Scholar]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef]
- Demirci, M.; Ünlü, Ö.; Tosun, A.I. Detection of O25b-ST131 clone, CTX-M-1 and CTX-M-15 genes via real-time PCR in Escherichia coli strains in patients with UTIs obtained from a university hospital in Istanbul. J. Infect. Public Health 2019, 12, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Madec, J.Y.; Haenni, M.; Métayer, V.; Saras, E.; Nicolas-Chanoine, M.H. High prevalence of the animal-associated blaCTX-M-1 IncI1/ST3 plasmid in human Escherichia coli isolates. Antimicrob. Agents Chemother. 2015, 59, 5860–5861. [Google Scholar] [CrossRef]
- Liao, X.-P.; Xia, J.; Yang, L.; Li, L.; Sun, J.; Liu, Y.-H.; Jiang, H.-X. Characterization of CTX-M-14-producing Escherichia coli from food-producing animals. Front. Microbiol. 2015, 6, 1136. [Google Scholar] [CrossRef]
- Valverde, A.; Cantón, R.; Garcillán-Barcia, M.P.; Novais, Â.; Galán, J.C.; Alvarado, A.; de la Cruz, F.; Baquero, F.; Coque, T.M. Spread of blaCTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob. Agents Chemother. 2009, 53, 5204–5212. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-F.; Zhang, W.-H.; Ren, S.-Q.; Yang, L.; Lü, D.-H.; Zeng, Z.-L.; Liu, Y.-H.; Jiang, H.-X. IncA/C Plasmid-Mediated Spread of CMY-2 in Multidrug-Resistant Escherichia coli from Food Animals in China. PLoS ONE 2014, 9, e96738. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Peirano, G.; Pitout, J.D.D. The Role of Epidemic Resistance Plasmids and International High-Risk Clones in the Spread of Multidrug-Resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef]
- Jamborova, I.; Johnston, B.D.; Papousek, I.; Kachlikova, K.; Micenkova, L.; Clabots, C.; Skalova, A.; Chudejova, K.; Dolejska, M.; Literak, I.; et al. Extensive Genetic Commonality among Wildlife, Wastewater, Community, and Nosocomial Isolates of Escherichia coli Sequence Type 131 (H30R1 and H30Rx Subclones) That Carry blaCTX-M-27 or blaCTX-M-15. Antimicrob. Agents Chemother. 2018, 62, e00519-18. [Google Scholar] [CrossRef] [PubMed]
- Brahmi, S.; Dunyach-Rémy, C.; Touati, A.; Lavigne, J.-P. CTX-M-15-producing Escherichia coli and the pandemic clone O25b-ST131 isolated from wild fish in Mediterranean Sea. Clin. Microbiol. Infect. 2015, 21, e18–e20. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Chen, X.; Lian, M.; Wen, F. Phenotypic and genotypic characterization of multi-drug-resistant Escherichia coli isolates harboring blaCTX-M group extended-spectrum β-lactamases recovered from pediatric patients in Shenzhen, southern China. Infect. Drug Resist. 2019, 12, 1325–1332. [Google Scholar] [CrossRef]
- Aibinu, I.; Odugbemi, T.; Koenig, W.; Ghebremedhin, B. Sequence Type ST131 and ST10 Complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin. Microbiol. Infect. 2012, 18, 2011–2013. [Google Scholar]
- McKinnon, J.; Chowdhury, P.R.; Djordjevic, S.P. Genomic analysis of multidrug-resistant Escherichia coli ST58 causing urosepsis. Int. J. Antimicrob. Agents 2018, 52, 430–435. [Google Scholar] [CrossRef]
- Aristizábal-Hoyos, A.M.; Rodríguez, E.A.; Arias, L.; Jiménez, J.N. High clonal diversity of multi-drug-resistant and extended spectrum β-lactamase-producing Escherichia coli in a wastewater treatment plant. J. Environ. Manag. 2019, 245, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Ibekwe, A.M.; Murinda, S.E.; Graves, A.K. Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. PLoS ONE 2011, 6, e20819. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Ewers, C.; Wieler, L.H. Extended-spectrum β-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2011, 2, 246. [Google Scholar] [CrossRef]
- Darwich, L.; Vidal, A.; Seminati, C.; Albamonte, A.; Casado, A.; López, F.; López, R.M.; Migura-Garcia, L. High prevalence and diversity of extended-spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS ONE 2019, 14, e0210686. [Google Scholar] [CrossRef] [PubMed]








| Continent | Number of Publications |
|---|---|
| Europe | 81 |
| South America | 21 |
| North America | 16 |
| Africa | 15 |
| Asia | 13 |
| Oceania | 2 |
| Central America | 1 |
| Species | Number of Publications | ||
|---|---|---|---|
| Mammals | Ungulates | Wild boar (Sus scrofa) | 9 |
| Roe deer (Capreolus capreolus) | 6 | ||
| Red deer (Cervus elaphus) | 4 | ||
| Mouflon (Ovis orientalis musimon) | 3 | ||
| Small mammals | Brown rat (Rattus norvegicus) | 5 | |
| Hedgehog (Erinaceus europaeus) | 3 | ||
| Black rat (Rattus rattus) | 3 | ||
| Carnivores | Red fox (Vulpes vulpes) | 5 | |
| Iberian wolf (Canis lupus signatus) | 2 | ||
| Badger (Meles meles) | 2 | ||
| Birds | Gulls | Herring gull (Larus argentatus) | 15 |
| Yellow-legged gull (Larus michahellis) | 12 | ||
| Lesser black-backed gull (Larus fuscus) | 6 | ||
| Black-backed gull (Chroicocephalus ridibundus) | 6 | ||
| Birds of prey | Black kites (Milvus migrans) | 8 | |
| Common buzzard (Buteo buteo) | 7 | ||
| Pigeons | Pigeon (Columba livia) | 8 | |
| Ducks | Duck (Anas platyrhynchos) | 6 | |
| Rooks | Rooks (Corvus frugilegus) | 6 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palmeira, J.D.; Cunha, M.V.; Carvalho, J.; Ferreira, H.; Fonseca, C.; Torres, R.T. Emergence and Spread of Cephalosporinases in Wildlife: A Review. Animals 2021, 11, 1765. https://doi.org/10.3390/ani11061765
Palmeira JD, Cunha MV, Carvalho J, Ferreira H, Fonseca C, Torres RT. Emergence and Spread of Cephalosporinases in Wildlife: A Review. Animals. 2021; 11(6):1765. https://doi.org/10.3390/ani11061765
Chicago/Turabian StylePalmeira, Josman D., Mónica V. Cunha, João Carvalho, Helena Ferreira, Carlos Fonseca, and Rita T. Torres. 2021. "Emergence and Spread of Cephalosporinases in Wildlife: A Review" Animals 11, no. 6: 1765. https://doi.org/10.3390/ani11061765
APA StylePalmeira, J. D., Cunha, M. V., Carvalho, J., Ferreira, H., Fonseca, C., & Torres, R. T. (2021). Emergence and Spread of Cephalosporinases in Wildlife: A Review. Animals, 11(6), 1765. https://doi.org/10.3390/ani11061765