Identification of Sex-Related Genes from the Three-Spot Swimming Crab Portunus sanguinolentus and Comparative Analysis with the Crucifix Crab Charybdis feriatus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptome Analysis and Validation
2.2. Analysis of Differentially Expressed Genes of P. sanguinolentus
2.3. RNA-Seq Results Verification
2.4. Comparison of the DEGs between P. sanguinolentus and C. feriatus
3. Results
3.1. DEGs of P. sanguinolentus
3.2. Comparison of Unigenes between P. sanguinolentus and C. feriatus
4. Discussion
4.1. DEG Profiles of P. sanguinolentus
4.2. Genes Differentially Expressed in Females and Males of P. sanguinolentus
4.3. Differences between the DEGs of P. sanguinolentus and C. feriatus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mantelattof, F.L.M.; Christofoletti, R. Naturalfeeding activity of the crab Callinectes ornatus (Portunidae) in Ubatuba Bay (São Paulo, Brazil): Influence of season, sex, size and molt stage. Mar. Biol. 2001, 138, 585–594. [Google Scholar] [CrossRef]
- Nevis, A.B.; Martinelli, J.M.; Carvalho, A.S.S.; Nahum, V.J.I. Abundance and spatial-temporal distribution of the family Portunidae (Crustacea, Decapoda) in the Curuçá estuary on the northern coast of Brazil. Braz. J. Aquatic. Sci. Technol. 2009, 13, 71–79. [Google Scholar] [CrossRef]
- Williams, M.J.; Primavera, J.H. Choosing tropical portunid species for culture, domestication and stock enhancement in the Indo-Pacific. Asian Fish. Sci. 2001, 14, 121–142. [Google Scholar]
- Waiho, K.; Fazhan, H.; Quinitio, E.T.; Baylon, J.C.; Fujaya, Y.; Azmie, G.; Wu, Q.; Shi, X.; Ikhwanuddin, M.; Ma, H. Larval rearing of mud crab (Scylla): What lies ahead. Aquaculture 2018, 493, 37–50. [Google Scholar] [CrossRef]
- Fazhan, H.; Waiho, K.; Azri, M.F.D.; Al-Hafiz, I.; Norfaizza, W.I.W.; Megat, F.H.; Jasmani, S.; Ma, H.; Ikhwanuddin, M. Sympatric occurrence and population dynamics of Scylla spp. in equatorial climate: Effects of rainfall, temperature and lunar phase. Estuar. Coast. Shelf Sci. 2017, 198, 299–310. [Google Scholar] [CrossRef]
- Apel, M.; Spiridonov, V.A. Taxonomy and zoogeography of the portunid crabs (Crustacea: Decapoda: Brachyura: Portunidae) of the Arabian Gulf and adjacent waters. Fauna Arabia 1998, 17, 159–331. [Google Scholar]
- Baylon, J.; Suzuki, H. Effects of changes in salinity and temperature on survival and development of larvae and juveniles of the crucifix crab Charybdis feriatus (Crustacea: Decapoda: Portunidae). Aquaculture 2007, 269, 390–401. [Google Scholar] [CrossRef]
- Padayatti, P.S. Notes on the population characteristics and reproductive biology of the portunid crab Charybdis feriatus (Linnaeus) at Cochin. Indian J. Fish. 1990, 37, 155–158. [Google Scholar]
- Jia, X.; Zhou, M.; Zou, Z.; Lin, P.; Wang, Y.; Zhang, Z. Identification and comparative analysis of the ovary and testis microRNAome of mud crab Scylla paramamosain. Mol. Reprod. Dev. 2018, 85, 519–531. [Google Scholar] [CrossRef]
- Yang, X.; Ikhwanuddin, M.; Li, X.; Lin, F.; Wu, Q.; Zhang, Y.; You, C.; Liu, W.; Cheng, Y.; Shi, X.; et al. Comparative transcriptome analysis provides insights into differentially expressed genes and long non-coding RNAs between ovary and testis of the mud crab (Scylla paramamosain). Mar. Biotechnol. 2018, 20, 20–34. [Google Scholar] [CrossRef]
- Meng, X.L.; Liu, P.; Jia, F.L.; Li, J.; Gao, B.Q. De novo Transcriptome analysis of Portunus trituberculatus ovary and testis by RNA-seq: Identification of genes involved in gonadal development. PLoS ONE 2015, 10, e0128659. [Google Scholar] [CrossRef] [Green Version]
- Lv, J.; Zhang, L.; Liu, P.; Li, J. Transcriptomic variation of eyestalk reveals the genes and biological processes associated with molting in Portunus trituberculatus. PLoS ONE 2017, 12, e0175315. [Google Scholar] [CrossRef] [Green Version]
- Li, E.; Wang, S.; Li, C.; Wang, X.; Chen, K.; Chen, L. Transcriptome sequencing revealed the genes and pathways involved in salinity stress of Chinese mitten crab, Eriocheir sinensis. Physiol. Genom. 2014, 46, 177–190. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Li, X.; Deng, Y.; Lu, Q.; Yang, Y.; Pan, J.; Ge, J.; Xu, Z. Comparative transcriptome sequencing of the hepatopancreas reveals differentially expressed genes in the precocious juvenile Chinese mitten crab, Eriocheir sinensis (Crustacea: Decapoda). Aquac. Res. 2016, 48, 3645–3656. [Google Scholar] [CrossRef]
- Sun, M.; Li, T.L.; Lee, S.C.; Wang, L. Transcriptome assembly and expression profiling of molecular responses to cadmium toxicity in hepatopancreas of the freshwater crab Sinopotamon henanense. Sci. Rep. 2016, 6, 19405. [Google Scholar] [CrossRef] [Green Version]
- Eldem, V.; Zararsiz, G.; Taşçi, T.; Duru, I.P.; Bakir, Y.; Erkan, M. Transcriptome Analysis for Non-Model Organism: Current Status and Best-Practices. In Applications of RNA-Seq and Omics Strategies-from Microorganisms to Human Health; IntechOpen: Rijeka, Croatia, 2017. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yan, H.F.; Wu, W.; Yu, H.; Ge, X.J. Comparative transcriptome analysis and marker development of two closely related Primrose species (Primula poissonii and Primula wilsonii). BMC Genom. 2013, 14, 329. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Miao, G.; Fazhan, H.; Waiho, K.; Zheng, H.; Li, S.; Ikhwanuddin, M.; Ma, H. Transcriptome-seq provides insights into sex-preference pattern of gene expression between testis and ovary of the crucifix crab (Charybdis feriatus). Physiol. Genom. 2018, 50, 393–405. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Miao, G.; Wu, Q.; Lin, F.; You, C.; Wang, S.; Aweya, J.J.; Ma, H. Transcriptome sequencing and molecular markers discovery in the gonads of Portunus sanguinolentus. Sci. Data 2018, 5, 180131. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Mantione, K.J.; Kream, R.M.; Kuzelova, H.; Ptacek, R.; Raboch, J.; Samuel, J.M.; Stefano, G.B. Comparing bioinformatics gene expression profiling methods: Microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 2014, 20, 138–142. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Wan, H.; Jiang, H.; Zhao, Y.; Zhang, X.; Hu, S.; Wang, Q. A transcriptome analysis of mitten crab testes (Eriocheir sinensis). Genet. Mol. Biol. 2011, 34, 136–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Wang, X.; Zou, Z.; Jia, X.; Wang, Y.; Zhang, Z. Transcriptome analysis of the differences in gene expression between testis and ovary in green mud crab (Scylla paramamosain). BMC Genom. 2014, 15, 585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, W.; Yang, Q.; Zhang, Y.; Ardavan, F.; Fang, H.; Zheng, H.; Li, S.; Zhang, Y.; Ikhwanuddin, M.; Ma, H. Integrative transcriptome sequencing reveals the molecular difference of maturation process of ovary and testis in mud crab Scylla paramamosain. Front. Mar. Sci. 2021, 8, 658091. [Google Scholar] [CrossRef]
- Farlora, R.; Araya-Garay, J.; Gallardo-Escárate, C. Discovery of sex-related genes through high-throughput transcriptome sequencing from the salmon louse Caligus rogercresseyi. Mar. Genom. 2014, 15, 85–93. [Google Scholar] [CrossRef]
- Decatur, W.A.; Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002, 27, 344–351. [Google Scholar] [CrossRef]
- Liu, Y.; Hui, M.; Cui, Z.; Luo, D.; Song, C.; Li, Y.; Liu, L. Comparative transcriptome analysis reveals sex-biased gene expression in juvenile Chinese mitten crab Eriocheir sinensis. PLoS ONE 2015, 10, e0133068. [Google Scholar] [CrossRef] [Green Version]
- Spaziani, E.P.; Hinsch, G.W.; Edwards, S.C. Changes in prostaglandin E2 and F2 alpha during vitellogenesis in the Florida crayfish Procambarus paeninsulanus. J. Comp. Physiol. B 1993, 163, 541–545. [Google Scholar] [CrossRef]
- Wimuttisuk, W.; Tobwor, P.; Deenarn, P.; Danwisetkanjana, K.; Pinkaew, D.; Kirtikara, K.; Vichai, V. Insights into the prostanoid pathway in the ovary development of the penaeid shrimp Penaeus monodon. PLoS ONE 2013, 8, e76934. [Google Scholar] [CrossRef] [Green Version]
- Warrier, S.; Subramoniam, T. Receptor mediated yolk protein uptake in the crab Scylla serrata: Crustacean vitellogenin receptor recognizes related mammalian serum lipoproteins. Mol. Reprod. Dev. 2002, 61, 536–548. [Google Scholar] [CrossRef]
- Duffy, D.M.; McGinnis, L.K.; VandeVoort, C.A.; Christenson, L.K. Mammalian oocytes are targets for prostaglandin E2 (PGE2) action. Reprod. Bio. Endocrinol. 2010, 8, 131. [Google Scholar] [CrossRef] [Green Version]
- Tahara, D.; Yano, I. Maturation-related variations in prostaglandin and fatty acid content of ovary in the kuruma prawn (Marsupenaeus japonicus). Comp. Biochem. Physiol. A 2004, 137, 631–637. [Google Scholar] [CrossRef]
- Kulkarni, G.K.; Nagabhushanam, R.; Joshi, P. Effect of progesterone on ovarian maturation in a marine penaeid prawn Parapenaeopsis hardwickii. Indian J. Exp. Biol. 1979, 17, 986–987. [Google Scholar]
- Coccia, E.; Lisa, E.D.; Cristo, C.D.; Cosmo, A.D.; Paolucci, M. Effects of estradiol and progesterone on the reproduction of the freshwater crayfish Cherax albidus. Biol. Bull. 2010, 218, 36–47. [Google Scholar] [CrossRef]
- Yang, F.; Xu, H.T.; Dai, Z.M.; Yang, W.J. Molecular characterization and expression analysis of vitellogenin in the marine crab Portunus trituberculatus. Comp. Biochem. Physiol. B 2005, 142, 456–464. [Google Scholar] [CrossRef]
- Mu, C.; Song, W.; Li, R.; Chen, Y.; Hao, G.; Wang, C. Identification of differentially expressed proteins relating to ovary development in Portunus trituberculatus. Aquaculture 2014, 426, 148–153. [Google Scholar] [CrossRef]
- Fliss, A.E.; Benzeno, S.; Rao, J.; Caplan, A.J. Control of estrogen receptor ligand binding by Hsp90. J. Steroid Biochem. Mol. Biol. 2000, 72, 223–230. [Google Scholar] [CrossRef]
- Wu, L.T.; Chu, K.H. Characterization of heat shock protein 90 in the shrimp Metapenaeusensis: Evidence for its role in the regulation of vitellogenin synthesis. Mol. Reprod. Dev. 2008, 75, 952–959. [Google Scholar] [CrossRef]
- Paolucci, M.; Di Cristo, C.; Di Cosmo, A. Immunological evidence for progesterone and estradiol receptors in the freshwater crayfish Austropotamobius pallipes. Mol. Reprod. Dev. 2002, 63, 55–62. [Google Scholar] [CrossRef]
- Luo, Y.; Amin, J.; Voellmy, R. Ecdysterone receptor is a sequence-specific transcription factor involved in the developmental regulation of heat shock genes. Mol. Cell Biol. 1991, 11, 3660–3675. [Google Scholar] [CrossRef] [Green Version]
- Özhan-Kizil, G.; Havermann, J.; Gerberding, M. Germ cells in the crustacean Parhyale hawaiensis depend on vasa protein for their maintenance but not for their formation. Dev. Biol. 2009, 327, 230–239. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, Y.; Han, K.; Zou, Z.; Zhang, Z. A vasa gene from green mud crab Scylla paramamosain and its expression during gonadal development and gametogenesis. Mol. Biol. Rep. 2012, 39, 4327–4335. [Google Scholar] [CrossRef]
- Aflalo, E.D.; Bakhrat, A.; Raviv, S.; Harari, D.; Sagi, A.; Abdu, U. Characterization of a vasa-like gene from the pacific white shrimp Litopenaeus vannamei and its expression during oogenesis. Mol. Reprod. Dev. 2007, 74, 172–177. [Google Scholar] [CrossRef]
- Simeó, C.S.; Andree, K.B.; Rotllant, G. Identification of vasa, a potential marker of primordial germ cells in the spider crab Maja brachydactyla, and its expression during early post-embryonic development. Invertebr. Reprod. Dev. 2011, 55, 91–99. [Google Scholar] [CrossRef]
- Fabioux, C.; Pouvreau, S.; Le Roux, F.; Huvet, A. The oyster vasa-like gene: A specific marker of the germline in Crassostrea gigas. Biochem. Biophys. Res. Commun. 2004, 315, 897–904. [Google Scholar] [CrossRef] [Green Version]
- Visudtiphole, V.; Klinbunga, S.; Kirtikara, K. Molecular characterization and expression profiles of cyclin A and cyclin B during ovarian development of the giant tiger shrimp Penaeus monodon. Comp. Biochem. Physiol. A 2009, 152, 535–543. [Google Scholar] [CrossRef]
- Han, K.; Dai, Y.; Zou, Z.; Fu, M.; Wang, Y.; Zhang, Z. Molecular characterization and expression profiles of cdc2 and cyclin B during oogenesis and spermatogenesis in green mud crab (Scylla paramamosain). Comp. Biochem. Physiol. B 2012, 163, 292–302. [Google Scholar] [CrossRef]
- Fang, J.J.; Qiu, G.F. Molecular cloning of cyclin B transcript with an unusually long 3′ untranslation region and its expression analysis during oogenesis in the Chinese mitten crab, Eriocheir sinensis. Mol. Biol. Rep. 2009, 36, 1521–1529. [Google Scholar] [CrossRef]
- Wu, P. Analysis of Ovary ESTs and cDNA Cloning and Expression Profile of Reproduction-Related Genes from Macrobrachium nipponense. Ph.D. Thesis, East China Normal University, Shanghai, China, 2009. [Google Scholar]
- Chang, C.C.; Tsai, K.W.; Hsiao, N.W.; Chang, C.Y.; Lin, C.L.; Watson, R.D.; Lee, C.Y. Structural and functional comparisons and production of recombinant crustacean hyperglycemic hormone (CHH) and CHH-like peptides from the mud crab Scylla olivacea. Gen. Comp. Endocrinol. 2010, 167, 68–76. [Google Scholar] [CrossRef]
- Song, C.; Cui, Z.; Hui, M.; Liu, Y.; Li, Y. Molecular characterization and expression profile of three Fem-1 genes in Eriocheir sinensis provide a new insight into crab sex-determining mechanism. Comp. Biochem. Physiol. B 2015, 189, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhu, D.; Yang, J.; Qiu, X.; Cui, X.; Tang, J. Molecular cloning of two structure variants of crustacean hyperglycemic hormone (CHH) from the swimming crab (Portunus trituberculatus), and their gene expression during molting and ovarian development. Zool. Sci. 2014, 31, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Chen, H.Y.; Choi, C.Y.; Roer, R.D.; Watson, R.D. Molecular cloning of a putative crustacean hyperglycemic hormone (CHH) isoform from extra-eyestalk tissue of the blue crab (Callinectes sapidus), and determination of temporal and spatial patterns of CHH gene expression. Gen. Comp. Endocrinol. 2010, 169, 174–181. [Google Scholar] [CrossRef]
- Vainio, S.; Heikkila, M.; Kispert, A.; Chin, N.; McMahon, A.P. Female development in mammals is regulated by Wnt-4 signalling. Nature 1999, 397, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Ardavan, F.; Fang, S.; Zhang, Y.; Cui, W.; Fang, H.; Ikhwanuddin, M.; Ma, H. The significant sex-biased expression pattern of Sp-Wnt4 provides novel insights into the ovarian development of mud crab (Scylla paramamosain). Int. J. Biol. Macromol. 2021, 183, 490–501. [Google Scholar] [CrossRef]
- Tevosian, S.G.; Manuylov, N.L. To β or not to β: Canonical β-catenin signaling pathway and ovarian development. Dev. Dynam. 2008, 237, 3672–3680. [Google Scholar] [CrossRef] [Green Version]
- Wilson, E.M.; French, F.S. Biochemical homology between rat dorsal prostate and coagulating gland. Purification of a major androgen-induced protein. J. Biol. Chem. 1980, 255, 10946–10953. [Google Scholar] [CrossRef]
- Leelatanawit, R.; Klinbunga, S.; Aoki, T.; Hirono, I.; Valyasevi, R.; Menasveta, P. Suppression subtractive hybridization (SSH) for isolation and characterization of genes related to testicular development in the giant tiger shrimp Penaeus monodon. J. Biochem. Mol. Biol. 2008, 41, 796–802. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Cui, Z. Molecular cloning and characterization of DMC1 from the Chinese mitten crab (Eriocheir sinensis). Int. J. Aquac. 2015, 1, 24–29. [Google Scholar]
- Qiao, H.; Xiong, Y.W.; Jiang, S.F.; Fu, H.T.; Sun, S.M.; Jin, S.B.; Gong, Y.S.; Zhang, W.Y. Gene expression profile analysis of testis and ovary of oriental river prawn Macrobrachium nipponense, reveals candidate reproduction-related genes. Genet. Mol. Res. 2015, 14, 2041–2054. [Google Scholar] [CrossRef]
- Manfrin, C.; Tom, M.; De Moro, G.; Gerdol, M.; Giulianini, P.G.; Pallavicini, A. The eyestalk transcriptome of red swamp crayfish Procambarus clarkii. Gene 2015, 557, 28–34. [Google Scholar] [CrossRef]
- Poels, J.; Van Loy, T.; Vandersmissen, H.P.; Van Hiel, B.; Van Soest, S.; Nachman, R.J.; Broeck, J.V. Myoinhibiting peptides are the ancestral ligands of the promiscuous Drosophila sex peptide receptor. Cell Mol. Life Sci. 2010, 67, 3511–3522. [Google Scholar] [CrossRef]
- Maurizio, O.; Liliana, S.M.; Romina, G.; Lorenza, Z.; Alessandro, N.; Maurizio, M.; Enrico, M.; Carlo, F. Evidence for FSH-dependent upregulation of SPATA2 (spermatogenesis-associated protein 2). Biochem. Biophy. Res. Commun. 2001, 283, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Sharabi, O.; Manor, R.; Weil, S.; Aflalo, E.D.; Lezer, Y.; Levy, T.; Mather, P.B.; Khalaila, I.; Sagi, A. Identification and characterization of an insulin-like receptor involved in crustacean reproduction. Endocrinology 2016, 157, 928–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Ye, H.; Huang, H.; Yang, Y.; Gong, J. An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen. Comp. Endocrinol. 2014, 204, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Bowles, J.; Schepers, G.; Koopman, P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000, 227, 239–255. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.Q.; Jiang, X.H.; Qi, H.Y.; Xiong, L.W.; Qiu, G.F. A novel SoxB2 gene is required for maturation of sperm nucleus during spermiogenesis in the Chinese mitten crab, Eriocheir sinensis. Sci. Rep. 2016, 6, 32139. [Google Scholar] [CrossRef] [Green Version]
- Murphy, M.W.; Zarkower, D.; Bardwell, V.J. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol. Biol. 2007, 8, 58. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.; Chen, Y.; Zou, Z.; Lin, P.; Wang, Y.; Zhang, Z. Characterization and expression profile of Vitellogenin gene from Scylla paramamosain. Gene 2013, 520, 119–130. [Google Scholar] [CrossRef]
- Kato, Y.; Kobayashi, K.; Watanabe, H.; Iguchi, T. Environmental sex determination in the branchiopod crustacean Daphnia magna: Deep conservation of a double sex gene in the sex-determining pathway. PLoS Genet. 2011, 7, e1001345. [Google Scholar] [CrossRef] [Green Version]
- Zarkower, D. Establishing sexual dimorphism: Conservation amidst diversity? Nat. Rev. Genet. 2001, 2, 175–185. [Google Scholar] [CrossRef]




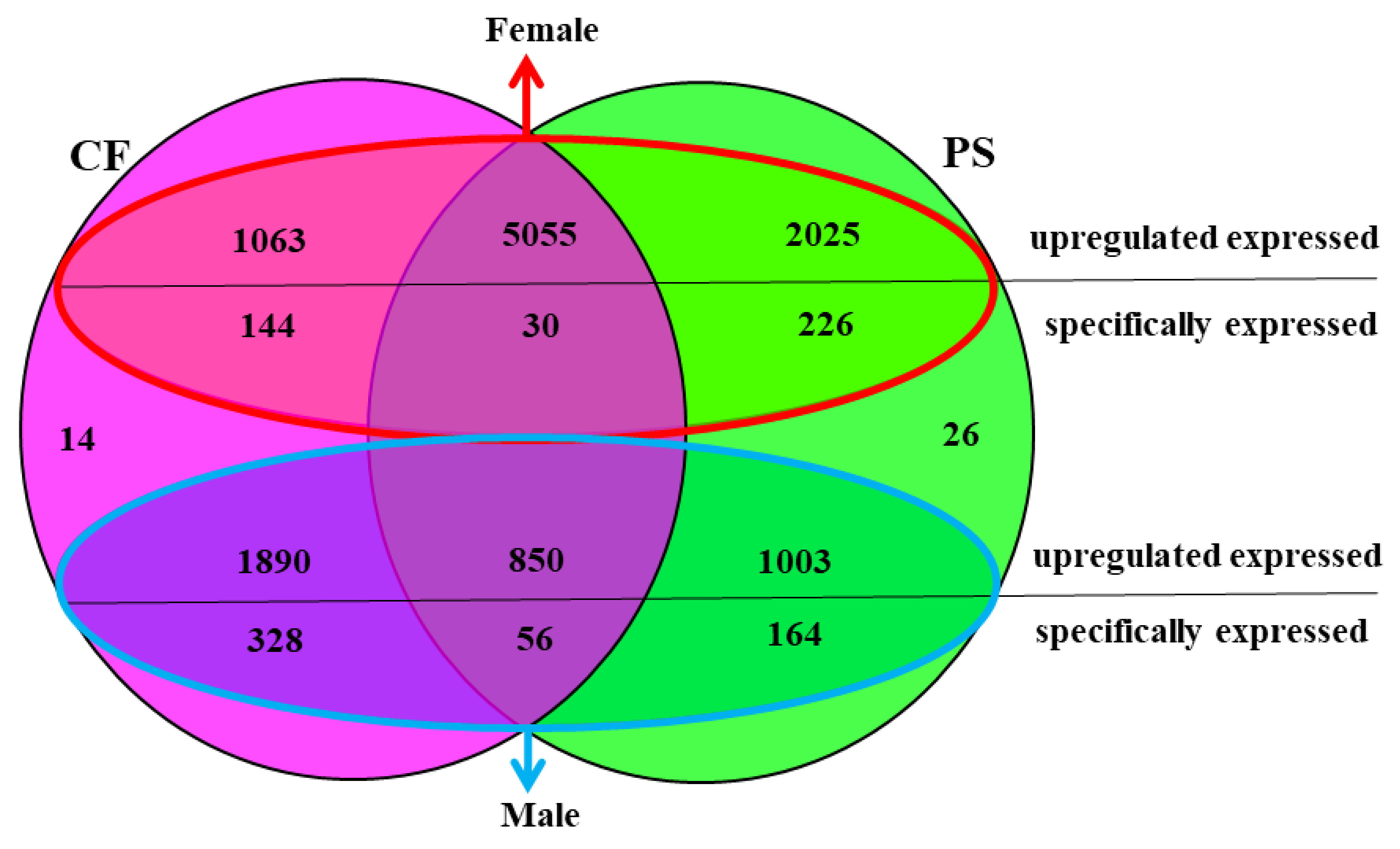
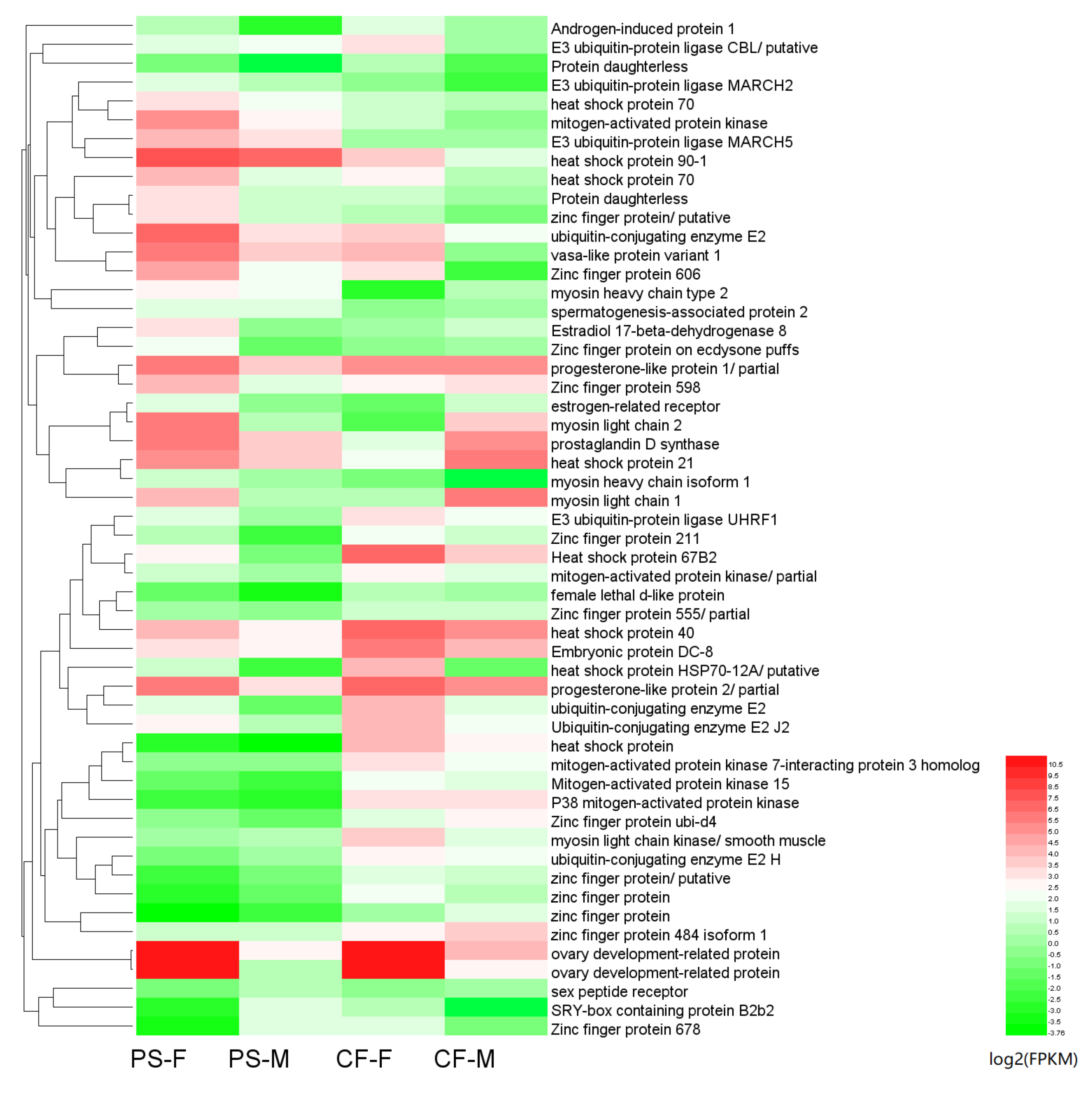

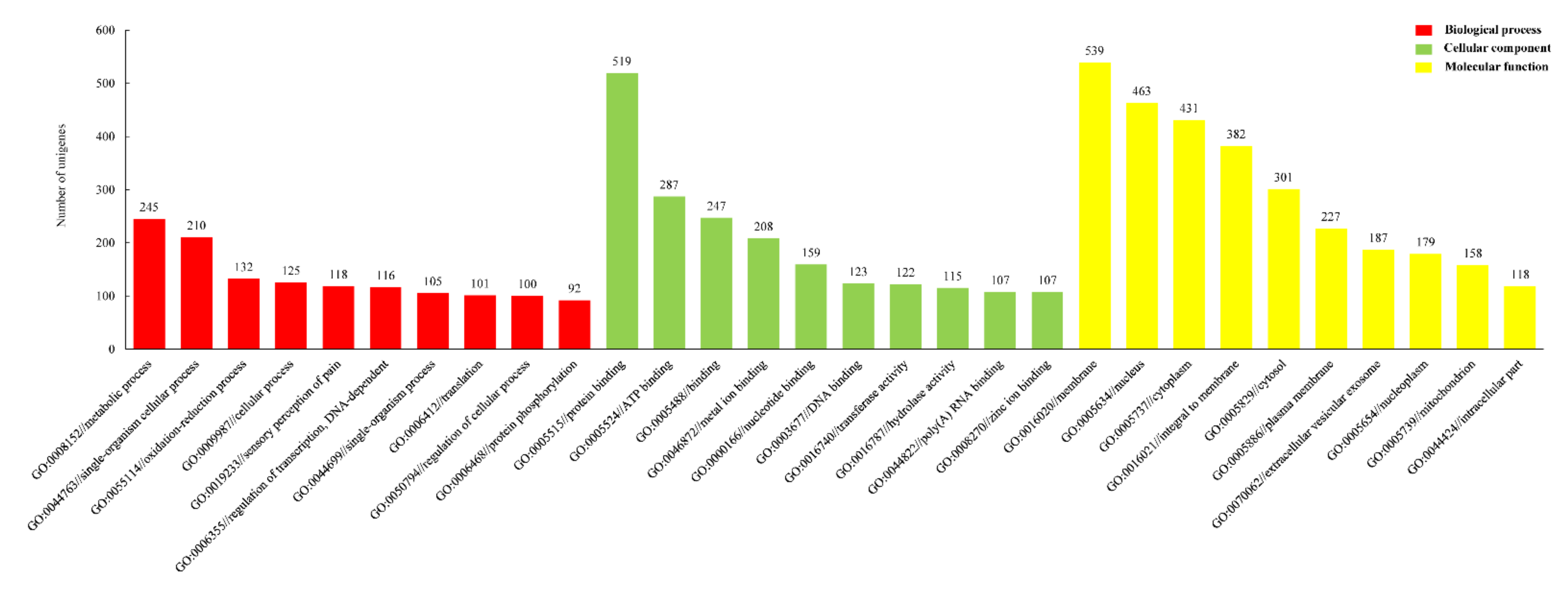
| FPKM | Gene Number of PS-F | Gene Number of PS-M | Gene Number of CF-F | Gene Number of CF-M |
|---|---|---|---|---|
| 0–100 | 119,142 | 119,469 | 85,851 | 86,074 |
| 101–1000 | 521 | 216 | 507 | 309 |
| 1001–10,000 | 47 | 23 | 63 | 45 |
| >10,000 | 8 | 10 | 12 | 5 |
| PS-M | PS-F | CF-M | CF-F | |
|---|---|---|---|---|
| Up-regulated | 1853 | 7080 | 2745 | 6118 |
| Specifically expressed | 220 | 256 | 384 | 174 |
| No expression | 26 | 14 | ||
| Total | 9435 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Waiho, K.; Ikhwanuddin, M.; Ma, H. Identification of Sex-Related Genes from the Three-Spot Swimming Crab Portunus sanguinolentus and Comparative Analysis with the Crucifix Crab Charybdis feriatus. Animals 2021, 11, 1946. https://doi.org/10.3390/ani11071946
Zhang Y, Waiho K, Ikhwanuddin M, Ma H. Identification of Sex-Related Genes from the Three-Spot Swimming Crab Portunus sanguinolentus and Comparative Analysis with the Crucifix Crab Charybdis feriatus. Animals. 2021; 11(7):1946. https://doi.org/10.3390/ani11071946
Chicago/Turabian StyleZhang, Yin, Khor Waiho, Mhd Ikhwanuddin, and Hongyu Ma. 2021. "Identification of Sex-Related Genes from the Three-Spot Swimming Crab Portunus sanguinolentus and Comparative Analysis with the Crucifix Crab Charybdis feriatus" Animals 11, no. 7: 1946. https://doi.org/10.3390/ani11071946






