Differentiating Pigs from Wild Boars Based on NR6A1 and MC1R Gene Polymorphisms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
Sampling
2.2. Methods
2.2.1. PCR Amplification, RFLP Genotyping and Sequencing of MC1R
2.2.2. Phylogenetic and Admixture Analysis
2.2.3. NR6A1 Gene Study and Real Time PCR
3. Results
3.1. MC1R
3.1.1. Sequence Study
3.1.2. RFLP Genotyping
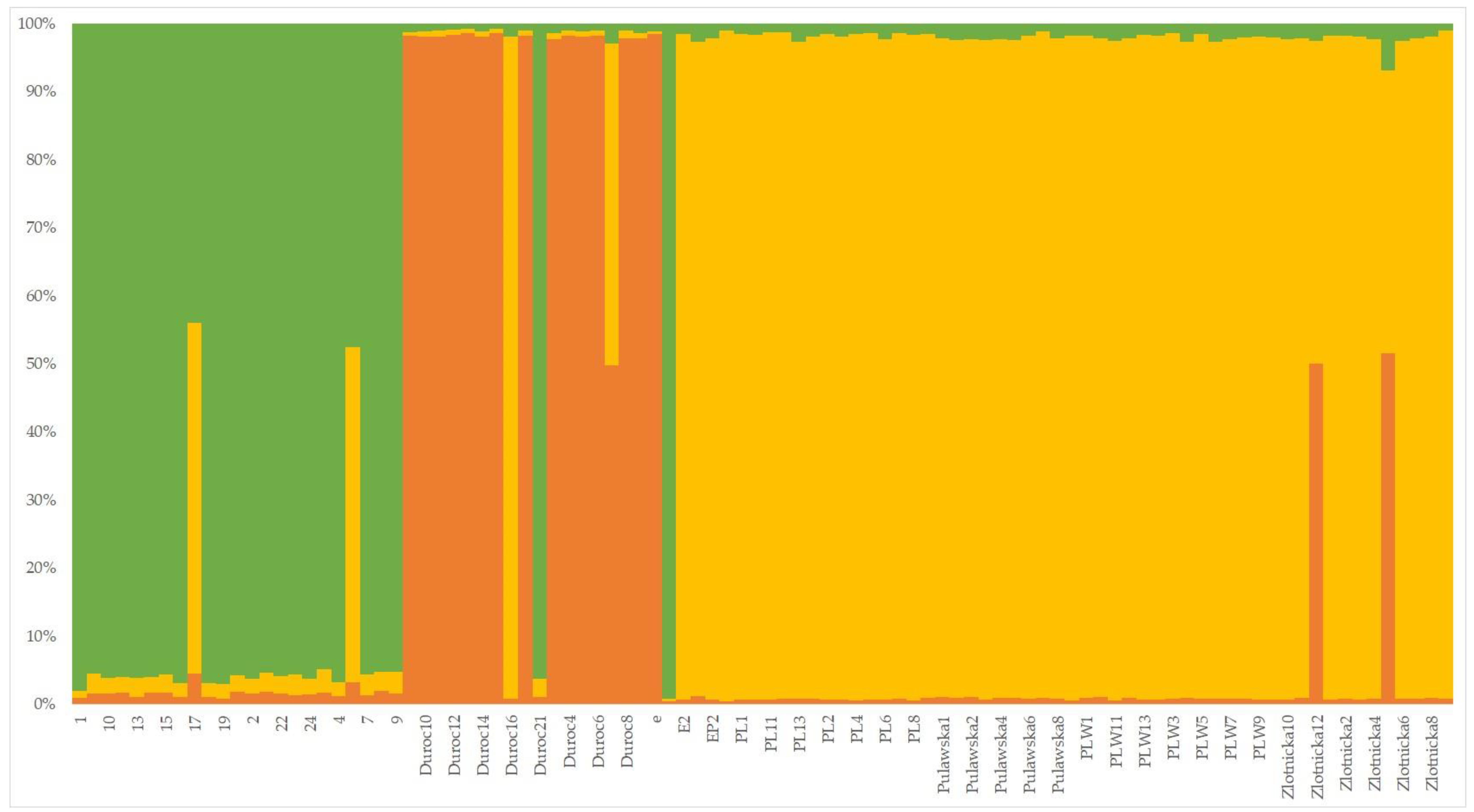
3.1.3. Phylogenetic and Population Characteristics of Wild Boars and Domestic Pigs
3.2. NR6A1 Allele Discrimination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Koutsogiannouli, E.A.; Moutou, K.A.; Sarafidou, T.; Stamatis, C.; Mamuris, Z. Detection of hybrids between wild boars (Sus scrofa scrofa) and domestic pigs (Sus scrofa f. domestica) in Greece, using the PCR-RFLP method on melanocortin-1 receptor (MC1R) mutations. Mamm. Biol. 2010, 75, 69–73. [Google Scholar] [CrossRef]
- Lorenzini, R.; Fanelli, R.; Tancredi, F.; Siclari, A.; Garofalo, L. Matching STR and SNP genotyping to discriminate between wild boar, domestic pigs and their recent hybrids for forensic purposes. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, D.C.; Lamoreux, M.L. The color loci of mice—A genetic century. Pigment Cell Res. 2003, 16, 333–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Willard, D.; Patel, I.R.; Kadwell, S.; Overton, L.; Kost, T.; Luther, M.; Chen, W.; Woychik, R.P.; Wilkison, W.O.; et al. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature 1994, 371, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Ollmann, M.M.; Lamoreux, M.L.; Wilson, B.D.; Barsh, G.S. Interaction of Agouti protein with the melanocortin 1 receptor in vitro and in vivo. Genes Dev. 1998, 12, 316–330. [Google Scholar] [CrossRef] [Green Version]
- Kijas, J.M.H.; Wales, R.; Törnsten, A.; Chardon, P.; Moller, M.; Andersson, L. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 1998, 150, 1177–1185. [Google Scholar] [CrossRef]
- Fang, M.; Larson, G.; Ribeiro, H.S.; Li, N.; Andersson, L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 2009, 5, e1000341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fajardo, V.; González, I.; Martín, I.; Rojas, M.; Hernández, P.E.; García, T.; Martín, R. Differentiation of European wild boar (Sus scrofa scrofa) and domestic swine (Sus scrofa domestica) meats by PCR analysis targeting the mitochondrial D-loop and the nuclear melanocortin receptor 1 (MC1R) genes. Meat Sci. 2008, 78, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Mikawa, S.; Morozumi, T.; Shimanuki, S.I.; Hayashi, T.; Uenishi, H.; Domukai, M.; Okumura, N.; Awata, T. Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1). Genome Res. 2007, 17, 586–593. [Google Scholar] [CrossRef] [Green Version]
- Ratajczak, M.; Buczyński, J.T. Origins and development of the Polish indigenous Złotnicka Spotted pig. Anim. Sci. Pap. Rep. 1997, 15, 137–147. [Google Scholar]
- Szyndler-Nędza, M.; Blicharski, T.; Bajda, Z. Świnie Puławskie—Uwarunkowania Kształtujące wielkość populacji w latach 1932–2007. Wiadomości Zootech. 2008, 4, 37–40. [Google Scholar]
- Janiszewski, P.; Grześkowiak, E.; Lisiak, D.; Szulc, K.; Borzuta, K. Quality and technological suitability of pork meat from Zlotnicka Spotted pigs and their crossbreeds with Duroc and Polish Large White. Sci. Ann. Pol. Soc. Anim. Prod. 2015, 11, 83–93. [Google Scholar]
- Hall, T.A. BioEdit, a user friendly biological sequence aligment editor.pdf. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Raymond, M.; Rousset, F. Genpop 1.2 Population genetics software for exact test and ecumenicism. J. Heredity 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Ogden, R.; Shuttleworth, C.; McEwing, R.; Cesarini, S. Median-joining networks for inferring intraspecific phylogenies. Conserv. Genet. 2005, 6, 37–48. [Google Scholar]
- Dzialuk, A.; Zastempowska, E.; Skórzewski, R.; Twarużek, M.; Grajewski, J. High domestic pig contribution to the local gene pool of free-living European wild boar: A case study in Poland. Mammal Res. 2018, 63, 65–71. [Google Scholar] [CrossRef] [Green Version]
- Babicz, M.; Pastwa, M.; Skrzypczak, E.; Buczyński, J.T. Variability in the melanocortin 1 receptor (MC1R) gene in wild boars and local pig breeds in Poland. Anim. Genet. 2013, 44, 357–358. [Google Scholar] [CrossRef] [PubMed]
- Russo, V.; Fontanesi, L.; Scotti, E.; Tazzoli, M.; Dall’Olio, S.; Davoli, R. Analysis of melanocortin 1 receptor (MC1R) gene polymorphisms in some cattle breeds: Their usefulness and application for breed traceability and authentication of Parmigiano Reggiano cheese. Ital. J. Anim. Sci. 2007, 6, 257–272. [Google Scholar] [CrossRef]
- D’Alessandro, E.; Fontanesi, L.; Liotta, L.; Davoli, R.; Chiofalo, V.; Russo, V. Analysis of the MC1R gene in the nero siciliano pig breed and usefulness of this locus for breed traceability. Vet. Res. Commun. 2007, 31, 389–392. [Google Scholar] [CrossRef]
- Fontanesi, L.; Ribani, A.; Scotti, E.; Utzeri, V.J.; Veličković, N.; Dall’Olio, S. Differentiation of meat from European wild boars and domestic pigs using polymorphisms in the MC1R and NR6A1 genes. Meat Sci. 2014, 98, 781–784. [Google Scholar] [CrossRef]
- Guzek, D.; Głąbska, D.; Głąbski, K.; Wierzbicka, A. Influence of Duroc breed inclusion into Polish landrace maternal line on pork meat quality traits. An. Acad. Bras. Cienc. 2016, 88, 1079–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canu, A.; Costa, S.; Iacolina, L.; Piatti, P.; Apollonio, M.; Scandura, M. Are captive wild boar more introgressed than free-ranging wild boar? Two case studies in Italy. Eur. J. Wildl. Res. 2014, 60, 459–467. [Google Scholar] [CrossRef]
- Luikart, G.; England, P.R.; Tallmon, D.; Jordan, S.; Taberlet, P. The power and promise of population genomics: From genotyping to genome typing. Nat. Rev. Genet. 2003, 4, 981–994. [Google Scholar] [CrossRef]
- Mikawa, S.; Hayashi, T.; Nii, M.; Shimanuki, S.; Morozumi, T.; Awata, T. Two quantitative trait loci on Sus scrofa chromosomes 1 and 7 affecting the number of vertebrae. J. Anim. Sci. 2005, 83, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer | Reference | Sequence Length (nt) | SNP Position * | RFLP/Real-Time PCR | Alleles (Band Length in bp) |
|---|---|---|---|---|---|---|
| MC1R | F: AGTGCCTGGAGGTGTCCATTCAC R: CGTAGATGAGGGGGTCCAGGATAGA | [8] | 575 | 370 | BspHI | G (672), A (472, 200) |
| 491 | - | CC, CT, TT | ||||
| 727 | BstUI | G (289, 222, 110), A (332, 289), | ||||
| 729 ** | - | A, G | ||||
| NR6A1 | F:AGGGCTTCAGAGAGCAACCA R:TGAAGCTCACCTGGAGGACAGT | [2] | 18 | 748 | VIC-TCACcGGGCTCCA-MGB NFQ FAM-CTCACtGGGCTCC-MGB NFQ | CC, CT, TT |
| MC1R Locus */Genotype ** and No. of Animals | MC1R Genotypes *** and No. of Animals | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| g.370G > A | g.491C > T | g.727G > A | ||||||||||||||
| Breed | GG | AG | AA | CC | CT | TT | GG | AG | AA | E+/E+ | E+/ED1,2,P2,3 | e/e | e/E D1,2,P2,3 | e/EP3 | ED1,2,P2/, ED1,2,P2,3 | EDD1,2,P2/EP3 |
| WB (n = 24) | 22 | 2 | - | 24 | - | - | 24 | - | - | 22 | 2 | - | - | - | - | - |
| D (n = 17) | 15 | 1 | 1 | 2 | 1 | 14 | 2 | 1 | 14 | 1 | 14 | 1 | 1 | - | ||
| ZW (n = 13) | - | 2 | 11 | 11 | 2 | - | 11 | 2 | - | - | - | 1 | 1 | 11 | - | |
| P (n = 10) | - | - | 10 | 10 | - | - | 10 | - | - | - | - | - | - | - | 10 | - |
| PL (n = 14) | - | - | 14 | 14 | - | - | 14 | - | - | - | - | - | 4 | 10 | ||
| PLW (n = 13) | - | - | 13 | 13 | - | - | 13 | - | - | - | - | - | 13 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koseniuk, A.; Smołucha, G.; Natonek-Wiśniewska, M.; Radko, A.; Rubiś, D. Differentiating Pigs from Wild Boars Based on NR6A1 and MC1R Gene Polymorphisms. Animals 2021, 11, 2123. https://doi.org/10.3390/ani11072123
Koseniuk A, Smołucha G, Natonek-Wiśniewska M, Radko A, Rubiś D. Differentiating Pigs from Wild Boars Based on NR6A1 and MC1R Gene Polymorphisms. Animals. 2021; 11(7):2123. https://doi.org/10.3390/ani11072123
Chicago/Turabian StyleKoseniuk, Anna, Grzegorz Smołucha, Małgorzata Natonek-Wiśniewska, Anna Radko, and Dominika Rubiś. 2021. "Differentiating Pigs from Wild Boars Based on NR6A1 and MC1R Gene Polymorphisms" Animals 11, no. 7: 2123. https://doi.org/10.3390/ani11072123
APA StyleKoseniuk, A., Smołucha, G., Natonek-Wiśniewska, M., Radko, A., & Rubiś, D. (2021). Differentiating Pigs from Wild Boars Based on NR6A1 and MC1R Gene Polymorphisms. Animals, 11(7), 2123. https://doi.org/10.3390/ani11072123






